Contents
- MAESpec_diffusion_fits.m
- Load perceptual learning data
- Design parameters
- Load diffusion data, MODEL 1 and MODEL 2
- Load diffusion data, MODEL 3
- Compare the two fits of MODEL 3
- Load diffusion data, MODEL 4
- Compare the two fits of MODEL 4
- Define Learning index (LI) and Specificity index (SI)
- Segment the data and calculate specificity indices
- This for loop is old (prior to Oct 2010). It is updated for 11 periods
- Descriptive statistics for the chi-squares for the saturated model
- G-squared statistic per subject for each of the 4 models:
- Fix the anomalous G-square values for sbj 363, MODEL 1
- Bayesian Information Criterion (BIC)
- BIC competition outcomes counted across the 27 subjects
- Specificity Indices (for all sbjs averaged together)
- dprime confidence interval calculations
- RT confidence interval calculations
- diffusion confidence interval calculations
- Plot RT and RT std (sbjs averaged together)
- Plot dprime
- All subjects plot with separate deltas
- Plot diffusion params (averaged across all sbjs)
- Export data for the six-plot empirical figure, 2010-11-17
- Raw specificity values
- Diffusion paramter specificity values (avg across subjs)
- Bootstrap Specificity Indices and Confidence Intervals
- B(1) contains the SIs and LIs on the full data set
- Average over all N_samples to find more stable SI values
- Average over all N_samples to find more stable LI values
- Collect distribution statistics of the 11-point profiles
- Plot bootstrap dprime curve and SI value for single dprime
- Bootstrap-based dprime learning curves
- Plot the resulting diffusion param bootstrap curves with SI written below
- Ter analysis
- New diffusion models (2-4)
- Plot Bootstrap RT and RT std (sbjs averaged together)
- Raw bootstrap percentiles
- Descriptive statistics for the on-line supplement, 2010-07-14
- Specificity-index summary table
- Learning-index summary table
- Bootstrap group-level Z-tests about specificity indices, accuracy
- Bootstrap group-level Z-tests about Learning indices, accuracy
- Bootstrap group-level Z-tests comparing the indices for Ter and MeanRT
- Individual-level Learning indices, accuracy and drift rates
- Paired-sample t-tests of LIs, individual-subject data
- Proportionality of easy and hard d'
- Within-sbj ANOVA for the boundary separation parameter a
- Define linear and quadratic trend coefficients
- Linear trend analysis of boundary separation parameter a
- Quadratic trend analysis of boundary separation parameter a
- Linear (and quadratic) trend analysis of drift-stdev parameter eta
- Linear (and quadratic) trend analysis of starting-point range parameter sz
- Linear (and quadratic) trend analysis of easy drift rate v1
- Linear (and quadratic) trend analysis of difficult drift rate v2
- Linear (and quadratic) trend analysis of the average drift rate v_avg
- Linear (and quadratic) trend analysis of the average dprime
- Linear (and quadratic) trend analysis of the easy dprime
- Linear (and quadratic) trend analysis of the hard dprime
- Linear (and quadratic) trend analysis of the mean raw RT (sRT_all)
- Linear (and quadratic) trend analysis of mean nondecision time Ter
- Linear (and quadratic) trend analysis of nondecision-time range parameter st
- Linear (and quadratic) trend analysis of minimum nondecision time t1
- Linear (and quadratic) trend analysis of maximum nondecision time t2
- Bootstrap group-level Z-tests about boundary separation
- Bootstrap group-level Z-tests about nondecision times
- Descriptive statistics of the drop of the maximum nondecision time t2
- Descriptive statistics of the drop of the mean RT
MAESpec_diffusion_fits.m
Plot results of the diffusion-model analysis of MAESpec experiments
File: work/MLExper/MAESpec02/diffusion/MAESpec_diffusion_fits.m
Usage: publish('MAESpec_diffusion_fits.m','html') ; %
'latex','doc','ppt'% The following files will be created and re-used. To start fresh, these % files must first be deleted manually: % .../work/MLExper/MAESpec02/diffusion/H.mat (hitcount results) % .../work/MLExper/MAESpec02/diffusion/B.mat (bootstrap results) % (c) Laboratory for Cognitive Modeling and Computational Cognitive % Neuroscience at the Ohio State University, http://cogmod.osu.edu % % 1.4.5 2010-11-23 AAP -- Descriptive stats for the chi2 for the saturated model % 1.4.4 2010-11-17 AAP -- Export data for 6-panel Figure and Table 1 in PB&R % 1.4.3 2010-11-10 AAP -- Trend analyses for dprime and raw RTs % 1.4.2 2010-11-06 AAP -- diffusion_params_par11terb and ..._par11terstb % 1.4.1 2010-10-27 AAP -- G-squared and Bayesian Info Criterion (BIC) % 1.4.0 2010-10-22 AAP -- Trend analyses % 1.3.0 2010-10-18 NVH -- Added 3 d models. Forked script into 11-period version % 1.2.0 2010-07-06 AAP -- More z-tests. Learning indices % 1.1.1 2010-04-15 AAP -- z-tests instead of t-tests at the end % 1.1.0 2010-04-14 NVH -- Now includes SIs for the RTs, etc. % 1.0.0 2010-03-24 NVH -- Initial version
Load perceptual learning data
Raw subject data is loaded from .../work/MLEXper/MAESpec01/data & .../work/MLExper/MAESpec02/data
As of 2010-10-21: The stuff in this section is old. It is about d' and mean RTs. It generates H.mat, which is now stable.
cd(fullfile(MAESpec02_pathstr,'diffusion')) ; fprintf('\n\nMAESpec_diffusion_fits executed on %s.\n\n',datestr(now)) ; fprintf('cd %s\n',pwd) ; clear all ; filename = fullfile(MAESpec02_pathstr,'diffusion','H.mat') ; %recalculatep = true ; recalculatep = false ; if (recalculatep || ~exist(filename,'file')) data_file = fullfile(MAESpec02_pathstr,'diffusion','raw_data.dat'); if (~exist(data_file, 'file')) file_1 = fullfile(MAESpec01_pathstr,'data','sbj*.dat'); file_2 = fullfile(MAESpec02_pathstr,'data','sbj*.dat'); %- Concatenate the individual data files into one master ASCII file fprintf('!cat %s %s > %s \n', file_1, file_2, data_file) ; eval(['!cat ' file_1 ' ' file_2 ' > ' data_file]); end %- Import to Matlab fprintf('\nD=MAES02_import_data ...') ; D=MAES02_import_data(fullfile(MAESpec02_pathstr,'diffusion','raw_data.dat')) ; %- Calculate d-primes fprintf('\nH = MAES02_hitcount(D) ...\n') ; H = MAES02_hitcount(D) ; save(filename,'H') ; fprintf('\nsave %s \n\n',filename) ; else fprintf('load %s \n\n',filename) ; load(filename) ; end N_sbj = length(H) ;
MAESpec_diffusion_fits executed on 29-Nov-2010 17:38:44. cd /Users/apetrov/a/r/w/work/MLExper/MAESpec02/diffusion load /Users/apetrov/a/r/w/work/MLExper/MAESpec02/diffusion/H.mat
Design parameters
These are fixed.
P = MAES02_params(0) ; design_params = P.design_params ; task_sched = design_params.task_sched ; % 1=MAE, 2=discrim N_sessions = design_params.N_session ; N_discrim_sessions = N_sessions(2) ; % 5 sessions N_MAE_sessions = N_sessions(1); % 2 sessions MAE_blocks_session = design_params.blocks_session(1) ; % 7 blocks per session discrim_blocks_session = design_params.blocks_session(2) ; % 8 blocks per session N_MAE_blocks = N_MAE_sessions * MAE_blocks_session ; N_discrim_blocks = N_discrim_sessions * discrim_blocks_session ; trials_MAE_block = design_params.trials_block(1) ; % 12 trials per block trials_discrim_block = design_params.trials_block(2) ;% 120 trials per block N_MAE_trials = N_MAE_blocks * trials_MAE_block ; N_discrim_trials = N_discrim_blocks * trials_discrim_block ;
Load diffusion data, MODEL 1 and MODEL 2
These data are described in .../work/MLExper/MAESpec02/diffusion/diffusion_params.txt
As of 2010-10-21, this is a fully saturated DM fit in which all params are allowed to vary for each period and each subject. One period is half-day's worth (4 blocks in MAESpec01_params terminology), except period 9, which is the "rogue block" immediately after the second MAE session. See blocks_per_period = [4 4 4 4 4 4 4 4 1 4 3] below. In later sections, this diffusion model fit is implicitly referred to as MODEL_1. Whenever there is something with no explicit subscript, it should be implicitly understood that the subscript is 1.
N_periods_total = 11 ; % as of 2010-10-18 df_1 = load(fullfile(MAESpec02_pathstr, 'diffusion', 'diffusion_params.txt'), '-ascii'); assert(all(size(df_1)==[N_sbj*N_periods_total 11])) ; % Load the new diffusion models (requested by reviewers) % As of 2010-10-21, these are MODEL_2, MODEL_3, and MODEL_4, as follows: % ** MODEL_2 is what Roger labeled 'par11a' in his email of 2010-10-09. % In MODEL_2, only drift is allowed to vary across periods. The same % boundary separation a and the same Ter and st apply across the board % for each individual subject. df_2 = load(fullfile(MAESpec02_pathstr, 'diffusion', 'diffusion_params_par11a.txt'), '-ascii'); assert(all(size(df_2)==[N_sbj 31])) ;
Load diffusion data, MODEL 3
** MODEL_3 is what Roger labeled 'par11tera' in his email of 2010-10-09. A second parameter search from a different starting point was performed on 2010-10-30 and emailed as 'par11terb'. In MODEL_3, both drift and Ter are allowed to vary across periods. The same boundary separation a and the same st apply across the board for each individual subject.
df_3a = load(fullfile(MAESpec02_pathstr, 'diffusion', 'diffusion_params_par11tera.txt'), '-ascii'); assert(all(size(df_3a)==[N_sbj 41])) ; df_3b = load(fullfile(MAESpec02_pathstr, 'diffusion', 'diffusion_params_par11terb.txt'), '-ascii'); assert(all(size(df_3b)==[N_sbj 40])) ; % Bring the two versions to a common data format df_3b = [df_3a(:,1) , df_3b] ; % append sbj_number as column 1 % The G-squared statistic (column 41) must be multiplied by 2 in par11tera % per Roger's email of 2010-10-27. The G^2 formula was corrected by the % time (2010-10-30) that par11terb was produced. % See Roger & Smith (2004, Psych Review, vol 111, equation on p. 343) df_3a(:,41) = df_3a(:,41).*2 ;
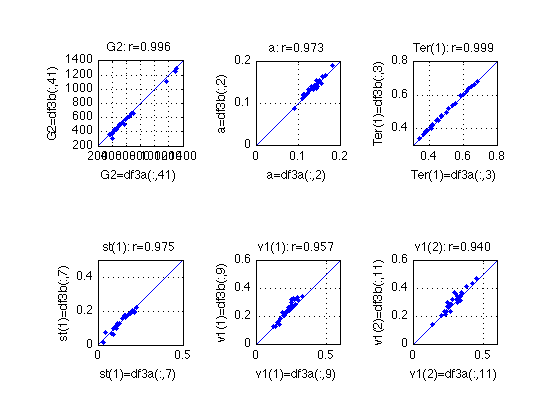
Compare the two fits of MODEL 3
idx = [41 2 3 7 9 11] ;
name = {'G2', 'a', 'Ter(1)', 'st(1)', 'v1(1)', 'v1(2)'} ;
ax = {[200 1400], [0 .2], [.300 .800], [0 .5], [0 .6], [0 .6]} ;
clf ;
for k = idx
kk = find(idx==k) ;
subplot(2,3,kk) ;
plot(df_3a(:,k),df_3b(:,k),'.');
axis([ax{kk} ax{kk}]) ; axis square ;
refline(1,0); grid on;
xlabel(sprintf('%s=df3a(:,%d)',name{kk},k));
ylabel(sprintf('%s=df3b(:,%d)',name{kk},k));
title(sprintf('%s: r=%.3f',name{kk},corr(df_3a(:,k),df_3b(:,k)))) ;
end
describe(df_3a(:,idx)-df_3b(:,idx),name) ;
% As the second run [df_3b=par11terb] yields lower G^2 values for all
% subjects, we'll adopt it for all subsequent calculations:
df_3 = df_3b ;
assert(all(df_3(:,41)<=df_3a(:,41))) ;
assert(all(df_3(:,41)<=df_3b(:,41))) ;
clear k name ax kk idx name
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
20.713 24.235 1.27 4.14 12.38 24.69 98.88 G2
-0.002 0.005 -0.01 -0.01 -0.00 -0.00 0.01 a
-0.003 0.005 -0.01 -0.01 -0.00 0.00 0.01 Ter(1)
0.000 0.012 -0.03 -0.01 -0.00 0.01 0.03 st(1)
-0.018 0.020 -0.07 -0.03 -0.01 -0.01 0.02 v1(1)
-0.015 0.025 -0.08 -0.02 -0.01 -0.00 0.03 v1(2)
------------------------------------------------------------
3.446 4.050 0.18 0.68 2.06 4.11 16.50
Load diffusion data, MODEL 4
** MODEL_4 is what Roger labeled 'par11tersta' in his email of 2010-10-09. A second parameter search from a different starting point was performed on 2010-10-30 and emailed as 'par11terstb'. In MODEL_4, three things (4 parameters total because there is easy and hard drift) are allowed to vary across periods. These are: - drift rate (easy and hard) - mean nondecision time Ter - range of the nondecision time distribution st The same boundary separation a applies across the board for each individual subject. Also the same eta and sz.
df_4a = load(fullfile(MAESpec02_pathstr, 'diffusion', 'diffusion_params_par11tersta.txt'), '-ascii'); assert(all(size(df_4a)==[N_sbj 51])) ; df_4b = load(fullfile(MAESpec02_pathstr, 'diffusion', 'diffusion_params_par11terstb.txt'), '-ascii'); assert(all(size(df_4b)==[N_sbj 50])) ; % Bring the two versions to a common data format df_4b = [df_4a(:,1) , df_4b] ; % append sbj_number as column 1 % The G-squared statistic (column 51) must be multiplied by 2 in par11tersta % per Roger's email of 2010-10-27. The G^2 formula was corrected by the % time (2010-10-30) that par11terstb was produced. % See Roger & Smith (2004, Psych Review, vol 111, equation on p. 343) df_4a(:,51) = df_4a(:,51).*2 ;
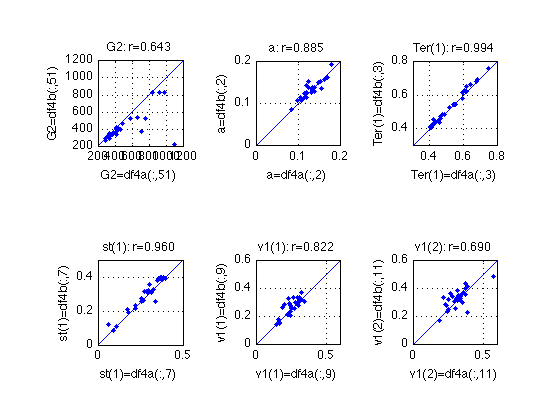
Compare the two fits of MODEL 4
idx = [51 2 3 7 9 11] ;
name = {'G2', 'a', 'Ter(1)', 'st(1)', 'v1(1)', 'v1(2)'} ;
ax = {[200 1200], [0 .2], [.300 .800], [0 .5], [0 .6], [0 .6]} ;
clf ;
for k = idx
kk = find(idx==k) ;
subplot(2,3,kk) ;
plot(df_4a(:,k),df_4b(:,k),'.');
axis([ax{kk} ax{kk}]) ; axis square ;
refline(1,0); grid on;
xlabel(sprintf('%s=df4a(:,%d)',name{kk},k));
ylabel(sprintf('%s=df4b(:,%d)',name{kk},k));
title(sprintf('%s: r=%.3f',name{kk},corr(df_4a(:,k),df_4b(:,k)))) ;
end
describe(df_4a(:,idx)-df_4b(:,idx),name) ;
% The fits aren't very stable. Among the two parameter sets for each
% subject, take the one that yields lower G^2. Note that there is no
% guarantee that this is the global minimum. In his email of 2010-10-30,
% Roger writes:
% "Some of the drift rates had not stabilized even after 7000 iterations.
% This was the largest number of drift rates fit in one program..."
df_4 = df_4b ;
idx = find(df_4(:,51)>df_4a(:,51)) ;
df_4(idx,:) = df_4a(idx,:) ;
df_4b(idx,[1 51]) % plot the subject numbers for whom the old fit was better
assert(all(df_4(:,51)<=df_4a(:,51))) ;
assert(all(df_4(:,51)<=df_4b(:,51))) ;
clear k name ax kk idx name
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
89.133 176.762 -16.16 15.03 34.61 82.42 883.65 G2
-0.002 0.010 -0.03 -0.01 0.00 0.00 0.02 a
-0.005 0.012 -0.03 -0.01 -0.00 0.00 0.02 Ter(1)
-0.013 0.027 -0.06 -0.03 -0.01 0.00 0.09 st(1)
-0.015 0.037 -0.10 -0.04 -0.01 0.01 0.04 v1(1)
-0.010 0.057 -0.12 -0.05 -0.01 0.02 0.16 v1(2)
------------------------------------------------------------
14.848 29.484 -2.75 2.48 5.76 13.74 147.33
ans =
356.0000 285.9980
362.0000 347.5690
Define Learning index (LI) and Specificity index (SI)
Input is a m-by-11 matrix of profiles. Output a m-by-1 vector of indices
assert(N_periods_total==11) ; % as of 2010-10-18 N_periods_train = 8 ; % 4 training sessions, each split in half %-- Learning index (LI) -- similar (but not identical) to that of Fine & Jacobs (2002) learn_idx = @(x) ((x(:,8)-x(:,1)) ./ x(:,1)) ; %-- Specificity index (SI) -- Ahissar & Hochstein (1997) specif_idx = @(x) ((x(:,8)-x(:,10)) ./ (x(:,8)-x(:,1))) ;
Segment the data and calculate specificity indices
These are all up-to-date as of 2010-10-21 -- all geared for 11 time periods
%[r,c] = recommend_subplot_size(N_sbj) ; r = 1; c = 2; M = 4.5 ; start_block = 4 ; end_block = 55 ; % Matrix of each subject's dprime and diffusion means for the 11 "blocks" of % interest % 1 row per sbj. 110 columns = dprime 1:11, then 11 columns per diffusion % parameter. Columns = % dprime; a; Ter; eta; sz; st; v1; v2 % followed by 11 columns of easy dprime and 11 columns of hard dprime sbj_means = NaN(N_sbj, 110) ; % G-squared goodness-of-fit statistic % See Roger & Smith (2004, Psych Review, vol 111, equation on p. 343) Gsq_1_per_period = NaN(N_sbj,N_periods_total) ; % G-squared for MODEL 1 % Matrix of specificty indices SI = NaN(N_sbj, N_periods_train) ; % using session 8 as the final day of training SIb = NaN(N_sbj, N_periods_train) ; % using session 8, block 1 as the final day of training SIc = NaN(N_sbj, N_periods_train) ; % using session 8, block 1 as the first day of *transfer* % Preallocate indv subject RT matrix sRT_all = NaN(N_sbj, N_periods_total) ; sRT_corr = NaN(N_sbj, N_periods_total) ; % RTs for correct trials only RTsd_all = NaN(N_sbj, N_periods_total) ; RTsd_corr = NaN(N_sbj, N_periods_total) ; %%%%%%%%%%%%%%%%%%%%%%% %%%%%%%%%%%%%%%%%%%%%%% % % Define indices so that so params can be called by name % These indices are relevant for MODEL_1 assert(N_periods_total==11) ; dpr = 1:11; a = 12:22; ter = 23:33; eta = 34:44; sz = 45:55; st = 56:66; v1 = 67:77; v2 = 78:88; edpr_idx = 89:99; hdpr_idx = 100:110; % 2010-10-20: The data layout of the various files that Roger emailed to us % over a period of several months is different. Thus, we need a separate % set of indices for each. % 2010-10-20: Define indices for the new diffusion MODELs 2 through 4. % These indices are relevant for MODEL_2 a_2 = 2 ; ter_2 = 3 ; eta_2 = 4 ; sz_2 = 5 ; st_2 = 7 ; v1_2 = 9:2:29 ; v2_2 = 10:2:30 ; gsq_2 = 31 ; % g squared value % These indices are relevant for MODEL_3 a_3 = 2 ; ter_3 = [3 31:40] ; eta_3 = 4 ; sz_3 = 5 ; st_3 = 7 ; v1_3 = 9:2:29 ; v2_3 = 10:2:30 ; gsq_3 = 41 ; % g squared value % These indices are relevant for MODEL_4 a_4 = 2 ; ter_4 = [3 31:40] ; eta_4 = 4 ; sz_4 = 5 ; st_4 = [7 41:50]; v1_4 = 9:2:29 ; v2_4 = 10:2:30 ; gsq_4 = 51 ; % g squared value
This for loop is old (prior to Oct 2010). It is updated for 11 periods
now (there used to be only 8 periods in the manuscript submitted to PBR on 2010-08-03). It is mostly about packaging, calculating means, and plotting d' and RTs.
for k=1:N_sbj % Average easy and hard dprime data into 11 periods like the % duffusion results. 4 blocks per period, except periods 9 & 11 dp11 = [mean(H(k).all_dprime(1:4))... % Session 1 (1/2) mean(H(k).all_dprime(5:8))... % Session 1 (2/2) mean(H(k).all_dprime(9:12))... % Session 2 (1/2) ... mean(H(k).all_dprime(13:16))... % Session 2 (2/2) ... mean(H(k).all_dprime(17:20))... % Session 3 (1/2) ... mean(H(k).all_dprime(21:24))... % Session 3 (2/2) mean(H(k).all_dprime(25:28))... % Session 4 (1/2) ... mean(H(k).all_dprime(29:32))... % Session 4 (2/2) mean(H(k).all_dprime(33))... % Session 5 *block 1* (1/3) mean(H(k).all_dprime(34:37))... % Session 5 (2/3) mean(H(k).all_dprime(38:40))]; % Session 5 (3/3) edp11 = [mean(H(k).easy_dprime(1:4))... % Session 1 (1/2) mean(H(k).easy_dprime(5:8))... % Session 1 (2/2) mean(H(k).easy_dprime(9:12))... % Session 2 (1/2) ... mean(H(k).easy_dprime(13:16))... % Session 2 (2/2) mean(H(k).easy_dprime(17:20))... % Session 3 (1/2) ... mean(H(k).easy_dprime(21:24))... % Session 3 (2/2) mean(H(k).easy_dprime(25:28))... % Session 4 (1/2) ... mean(H(k).easy_dprime(29:32))... % Session 4 (2/2) mean(H(k).easy_dprime(33))... % Session 5 *block 1* (1/3) mean(H(k).easy_dprime(34:37))... % Session 5 (2/3) mean(H(k).easy_dprime(38:40))]; % Session 5 (3/3) hdp11 = [mean(H(k).hard_dprime(1:4))... % Session 1 (1/2) mean(H(k).hard_dprime(5:8))... % Session 1 (2/2) mean(H(k).hard_dprime(9:12))... % Session 2 (1/2) ... mean(H(k).hard_dprime(13:16))... % Session 2 (2/2) mean(H(k).hard_dprime(17:20))... % Session 3 (1/2) ... mean(H(k).hard_dprime(21:24))... % Session 3 (2/2) mean(H(k).hard_dprime(25:28))... % Session 4 (1/2) ... mean(H(k).hard_dprime(29:32))... % Session 4 (2/2) mean(H(k).hard_dprime(33))... % Session 5 *block 1* (1/3) mean(H(k).hard_dprime(34:37))... % Session 5 (2/3) mean(H(k).hard_dprime(38:40))]; % Session 5 (3/3) % Collect RT statistics % Mean RT (of *median* block RTs) sRT_all(k,:) = [mean(H(k).RT_descr(1:4,5),1)... % Session 1 (1/2) mean(H(k).RT_descr(5:8,5),1)... % Session 1 (2/2) mean(H(k).RT_descr(9:12,5),1)... % Session 2 (1/2) ... mean(H(k).RT_descr(13:16,5),1)... % Session 2 (2/2) mean(H(k).RT_descr(17:20,5),1)... % Session 3 (1/2) ... mean(H(k).RT_descr(21:24,5),1)... % Session 3 (2/2) mean(H(k).RT_descr(25:28,5),1)... % Session 4 (1/2) ... mean(H(k).RT_descr(29:32,5),1)... % Session 4 (2/2) mean(H(k).RT_descr(33,5),1)... % Session 5 *block 1* (1/3) mean(H(k).RT_descr(34:37,5),1)... % Session 5 (2/3) mean(H(k).RT_descr(38:40,5),1)]; % Session 5 (3/3) sRT_corr(k,:) = [mean(H(k).RT_descr_corr(1:4,5),1)... % Session 1 (1/2) mean(H(k).RT_descr_corr(5:8,5),1)... % Session 1 (2/2) mean(H(k).RT_descr_corr(9:12,5),1)... % Session 2 (1/2) ... mean(H(k).RT_descr_corr(13:16,5),1)... % Session 2 (2/2) mean(H(k).RT_descr_corr(17:20,5),1)... % Session 3 (1/2) ... mean(H(k).RT_descr_corr(21:24,5),1)... % Session 3 (2/2) mean(H(k).RT_descr_corr(25:28,5),1)... % Session 4 (1/2) ... mean(H(k).RT_descr_corr(29:32,5),1)... % Session 4 (2/2) mean(H(k).RT_descr_corr(33,5),1)... % Session 5 *block 1* (1/3) mean(H(k).RT_descr_corr(34:37,5),1)... % Session 5 (2/3) mean(H(k).RT_descr_corr(38:40,5),1)]; % Session 5 (3/3) % std of RT across "blocks" RTsd_all(k,:) = [mean(H(k).RT_descr(1:4,2),1)... % Session 1 (1/2) mean(H(k).RT_descr(5:8,2),1)... % Session 1 (2/2) mean(H(k).RT_descr(9:12,2),1)... % Session 2 (1/2) ... mean(H(k).RT_descr(13:16,2),1)... % Session 2 (2/2) mean(H(k).RT_descr(17:20,2),1)... % Session 3 (1/2) ... mean(H(k).RT_descr(21:24,2),1)... % Session 3 (2/2) mean(H(k).RT_descr(25:28,2),1)... % Session 4 (1/2) ... mean(H(k).RT_descr(29:32,2),1)... % Session 4 (2/2) mean(H(k).RT_descr(33,2),1)... % Session 5 *block 1* (1/3) mean(H(k).RT_descr(34:37,2),1)... % Session 5 (2/3) mean(H(k).RT_descr(38:40,2),1)]; % Session 5 (3/3) RTsd_corr(k,:) = [mean(H(k).RT_descr_corr(1:4,2),1)... % Session 1 (1/2) mean(H(k).RT_descr_corr(5:8,2),1)... % Session 1 (2/2) mean(H(k).RT_descr_corr(9:12,2),1)... % Session 2 (1/2) ... mean(H(k).RT_descr_corr(13:16,2),1)... % Session 2 (2/2) mean(H(k).RT_descr_corr(17:20,2),1)... % Session 3 (1/2) ... mean(H(k).RT_descr_corr(21:24,2),1)... % Session 3 (2/2) mean(H(k).RT_descr_corr(25:28,2),1)... % Session 4 (1/2) ... mean(H(k).RT_descr_corr(29:32,2),1)... % Session 4 (2/2) mean(H(k).RT_descr_corr(33,2),1)... % Session 5 *block 1* (1/3) mean(H(k).RT_descr_corr(34:37,2),1)... % Session 5 (2/3) mean(H(k).RT_descr_corr(38:40,2),1)]; % Session 5 (3/3) % Store sbj dprime means sbj_means(k, dpr) = dp11 ; % Isolate diffusion params for the current subject idx = find(df_1(:,1)==H(k).sbj) ; % sanity check current_sbj = H(k).sbj ; assert(all(df_1(idx,1)==current_sbj)) ; assert(df_2(k,1)==current_sbj) ; assert(df_2(k,1)==current_sbj) ; assert(df_2(k,1)==current_sbj) ; % param "a" sbj_means(k,a) = df_1(idx, 2)'; % param "Ter" sbj_means(k,ter) = df_1(idx, 3)'; % param "eta" sbj_means(k,eta) = df_1(idx, 4)'; % param "sz" sbj_means(k,sz) = df_1(idx, 5)'; % param "st" sbj_means(k,st) = df_1(idx, 7)'; % param "v1" sbj_means(k,v1) = -df_1(idx, 9)'; % param "v2" sbj_means(k,v2) = -df_1(idx, 10)'; % G-squared goodness-of-fit statistic Gsq_1_per_period(k,:) = df_1(idx,11)' ; % easy dprime sbj_means(k,edpr_idx) = edp11 ; % difficult dprime sbj_means(k,hdpr_idx) = hdp11 ; % Easy/Hard specificity indices for plotting %e_SI = (edp8(5) - edp8(7)) / (edp8(5) - edp8(1)) ; %h_SI = (hdp8(5) - hdp8(7)) / (hdp8(5) - hdp8(1)) ; e_SI = specif_idx(edp11) ; h_SI = specif_idx(hdp11) ; % Specificity Indices using session 4 as final "block" of training data SI(k, :) = [ (sbj_means(k,dpr(8))-sbj_means(k,dpr(10)))/(sbj_means(k,dpr(8))-sbj_means(k,dpr(1))),... (sbj_means(k,a(8))-sbj_means(k,a(10)))/(sbj_means(k,a(8))-sbj_means(k,a(1))),... (sbj_means(k,ter(8))-sbj_means(k,ter(10)))/(sbj_means(k,ter(8))-sbj_means(k,ter(1))),... (sbj_means(k,eta(8))-sbj_means(k,eta(10)))/(sbj_means(k,eta(8))-sbj_means(k,eta(1))),... (sbj_means(k,sz(8))-sbj_means(k,sz(10)))/(sbj_means(k,sz(8))-sbj_means(k,sz(1))),... (sbj_means(k,st(8))-sbj_means(k,st(10)))/(sbj_means(k,st(8))-sbj_means(k,st(1))),... (sbj_means(k,v1(8))-sbj_means(k,v1(10)))/(sbj_means(k,v1(8))-sbj_means(k,v1(1))),... (sbj_means(k,v2(8))-sbj_means(k,v2(10)))/(sbj_means(k,v2(8))-sbj_means(k,v2(1)))] ; SIb(k, :) = [ (sbj_means(k,dpr(9))-sbj_means(k,dpr(10)))/(sbj_means(k,dpr(9))-sbj_means(k,dpr(1))),... (sbj_means(k,a(9))-sbj_means(k,a(10)))/(sbj_means(k,a(9))-sbj_means(k,a(1))),... (sbj_means(k,ter(9))-sbj_means(k,ter(10)))/(sbj_means(k,ter(9))-sbj_means(k,ter(1))),... (sbj_means(k,eta(9))-sbj_means(k,eta(10)))/(sbj_means(k,eta(9))-sbj_means(k,eta(1))),... (sbj_means(k,sz(9))-sbj_means(k,sz(10)))/(sbj_means(k,sz(9))-sbj_means(k,sz(1))),... (sbj_means(k,st(9))-sbj_means(k,st(10)))/(sbj_means(k,st(9))-sbj_means(k,st(1))),... (sbj_means(k,v1(9))-sbj_means(k,v1(10)))/(sbj_means(k,v1(9))-sbj_means(k,v1(1))),... (sbj_means(k,v2(9))-sbj_means(k,v2(10)))/(sbj_means(k,v2(9))-sbj_means(k,v2(1)))] ; SIc(k, :) = [ (sbj_means(k,dpr(8))-sbj_means(k,dpr(9)))/(sbj_means(k,dpr(8))-sbj_means(k,dpr(1))),... (sbj_means(k,a(8))-sbj_means(k,a(9)))/(sbj_means(k,a(8))-sbj_means(k,a(1))),... (sbj_means(k,ter(8))-sbj_means(k,ter(9)))/(sbj_means(k,ter(8))-sbj_means(k,ter(1))),... (sbj_means(k,eta(8))-sbj_means(k,eta(9)))/(sbj_means(k,eta(8))-sbj_means(k,eta(1))),... (sbj_means(k,sz(8))-sbj_means(k,sz(9)))/(sbj_means(k,sz(8))-sbj_means(k,sz(1))),... (sbj_means(k,st(8))-sbj_means(k,st(9)))/(sbj_means(k,st(8))-sbj_means(k,st(1))),... (sbj_means(k,v1(8))-sbj_means(k,v1(9)))/(sbj_means(k,v1(8))-sbj_means(k,v1(1))),... (sbj_means(k,v2(8))-sbj_means(k,v2(9)))/(sbj_means(k,v2(8))-sbj_means(k,v2(1)))] ; % % Plot dprime % subplot(r,c,1) ; % plot(1:11, edp11,'b.-') ; % hold on % plot(1:11, hdp11, 'r.-') ; % axis([1 11 0 M]) ; grid on ; % set(gca,'xtick',([1:11]),... % 'xticklabel',[],'ytick',(0:ceil(M))) ; % title(sprintf('Sbj %d, gr %d',H(k).sbj,H(k).group)) ; % ylabel('dprime'); % xlabel(sprintf('SI(e) = %0.3f ; SI(h) = %0.3f', e_SI, h_SI)); % hold off % % % Plot drift rates % subplot(r,c,2) ; % plot(1:11, sbj_means(k,v1), 'b.-'); % hold on % plot(1:11, sbj_means(k,v2), 'r.-'); % axis([1 11 0 1]) ; grid on ; % set(gca,'xtick',([1:11]),... % 'xticklabel',[],'ytick',(0:.1:1)) ; % title(sprintf('Sbj %d, gr %d',H(k).sbj,H(k).group)) ; % ylabel('drift rate'); % xlabel(sprintf('SI(v1) = %0.3f ; SI(v2) = %0.3f', SI(k,7), SI(k,8))); % legend('v1','v2','location','NorthEast'); % hold off end
Descriptive statistics for the chi-squares for the saturated model
Added by Alex, 2010-11-23:
When Roger did his original fit (MODEL 1), he used ordinary chi-square to evaluate the goodness of fit. Then he switched to Gsq for Models 2, 3, and 4. The motivation for this was that Gsq is the mathematically appropriate term for the BIC calculation. Both are asymptotically distributed as chi-square with the same df. However, MODEL 1 (the saturated model) was never re-fit to optimize Gsq instead of chi2.
This is just as well because in the final manuscript we decided to stick to Model 1 (given that most parameters had statistically significant learning trends) and to report the standard chi-squared measure of fit.
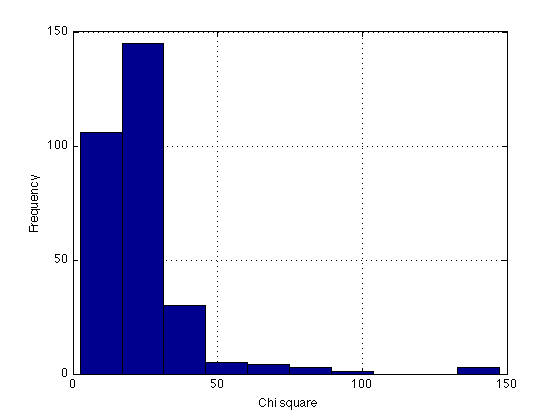
%- Goodness-of-fit measure for the diffusion model % Chi-square values, as per Roger's email of 2010-01-19. % The 6-parameter DDM (z=a/2) was fit to individual data in each period, % after which the parameter values (and the chi-squares?) were averaged % across Ss to yield the following: chi2=[38.185 27.154 27.034 22.644 24.842 15.799 18.845 19.900] ; fprintf('\n Chi2 per Roger''s email: summed across blocks:') ; describe(chi2,'chi2') % This is close to the critical chi2=25.0=chi2inv(.95,15) % df=15, alpha=5% % This indicates excellent fits -- most of the discrepancy between model % and data can be attributed to sampling fluctuations. critical_chi2 = chi2inv(.95,15) %- Calculated from the imported DM fits: summed across Ss, varies across blocks block_legend = cell(N_periods_total,1) ; fprintf('\n Chi2 for saturated model: summed across Ss, varies across blocks:') ; for k=1:N_periods_total ; block_legend{k} = sprintf('Block %2d',k) ; end describe(Gsq_1_per_period,block_legend) ; % summed across all Ss %- Calculated from the imported DM fits: summed across blocks, varies across Ss sbj_legend = cell(N_sbj,1) ; fprintf('\n Chi2 for saturated model: summed across blocks, varies across Ss:') ; for k=1:N_sbj ; sbj_legend{k} = sprintf('Sbj %3d',H(k).sbj) ; end describe(Gsq_1_per_period',sbj_legend) ; % summed across all blocks %- Summed across everything fprintf('\n Chi2 for saturated model: summed across both Ss and blocks:') ; describe(Gsq_1_per_period(:)) ; clf ; hist(Gsq_1_per_period(:)) ; grid on xlabel('Chi square') ; ylabel('Frequency') ; % Number of significant deviations k = Gsq_1_per_period>critical_chi2 ; % [N_sbj x N_periods_total] fprintf('\n Number of significant chi2 in each block:\n %s \n' ... ,mat2str(sum(k))) ; fprintf('\n Number of significant chi2 in each subject:\n %s \n' ... ,mat2str(sum(k,2)')) ; fprintf('\n Total number of significant chi2: %d out of %d \n'... ,sum(k(:)), length(k(:))) ; % Number of large deviations acceptable_chi2 = 2*critical_chi2 k = Gsq_1_per_period>acceptable_chi2 ; % [N_sbj x N_periods_total] fprintf('\n Number of unacceptable chi2 in each block:\n %s \n' ... ,mat2str(sum(k))) ; fprintf('\n Number of unacceptable chi2 in each subject:\n %s \n' ... ,mat2str(sum(k,2)')) ; fprintf('\n Total number of unacceptable chi2: %d out of %d \n'... ,sum(k(:)), length(k(:))) ; k = sort(Gsq_1_per_period(:),'descend')' ; fprintf('\n The ten most deviating values:\n %s \n', mat2str(k(1:10))) ; clear k ;
Chi2 per Roger's email: summed across blocks:
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
24.300 6.895 15.80 19.37 23.74 27.09 38.19 chi2
critical_chi2 =
24.9958
Chi2 for saturated model: summed across Ss, varies across blocks:
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
36.115 34.193 10.63 18.05 22.46 30.38 141.71 Block 1
26.342 10.846 9.96 18.63 23.07 33.94 48.92 Block 2
23.645 9.965 9.22 15.46 21.83 30.17 52.82 Block 3
29.842 30.234 6.42 16.13 20.48 29.43 147.43 Block 4
23.272 12.604 9.90 16.80 21.78 26.81 77.81 Block 5
23.610 13.615 3.27 14.11 21.48 30.74 70.72 Block 6
19.757 7.572 8.24 14.28 18.38 23.59 38.66 Block 7
24.772 15.879 4.54 13.94 20.51 31.76 72.70 Block 8
14.760 5.833 4.94 10.44 13.89 20.55 23.91 Block 9
18.378 8.444 2.46 13.08 15.89 22.81 37.08 Block 10
19.295 7.834 5.58 14.50 18.59 22.10 40.73 Block 11
------------------------------------------------------------
23.617 14.274 6.83 15.04 19.85 27.48 68.41
Chi2 for saturated model: summed across blocks, varies across Ss:
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
24.320 11.509 10.91 15.43 21.40 33.28 46.90 Sbj 353
18.705 10.997 4.54 9.71 16.60 26.54 39.62 Sbj 354
21.325 7.710 11.43 15.75 19.04 27.60 35.27 Sbj 355
19.514 6.806 12.05 13.79 17.14 26.48 30.55 Sbj 356
25.630 21.531 11.19 14.10 18.56 21.89 86.32 Sbj 357
19.198 8.293 2.46 15.03 19.68 22.20 33.89 Sbj 359
40.724 40.957 9.22 14.22 28.24 45.88 141.71 Sbj 360
22.261 10.735 10.42 14.49 19.42 27.64 41.39 Sbj 362
34.154 28.968 4.94 14.37 20.41 62.80 81.65 Sbj 363
21.709 8.907 7.24 16.96 21.41 30.33 34.68 Sbj 364
17.953 6.609 6.42 13.53 18.22 21.49 28.48 Sbj 366
21.412 7.116 10.97 14.52 23.41 27.59 29.74 Sbj 367
24.616 6.135 15.50 21.10 22.98 28.88 36.05 Sbj 382
18.997 8.426 9.49 12.21 17.05 24.50 35.10 Sbj 383
16.155 4.581 6.68 14.22 14.89 19.98 23.23 Sbj 384
27.555 9.524 15.14 19.57 25.55 33.98 45.02 Sbj 385
18.793 5.862 9.11 15.14 18.20 24.48 26.23 Sbj 386
32.301 38.845 10.64 14.35 20.53 28.28 147.43 Sbj 387
21.584 9.534 9.71 12.47 22.09 25.71 42.10 Sbj 388
19.541 8.291 9.96 14.06 18.46 23.47 37.08 Sbj 389
30.981 22.691 8.59 16.49 20.73 51.28 70.72 Sbj 390
32.625 34.764 11.41 18.07 24.49 29.29 135.67 Sbj 391
28.329 11.476 17.14 18.83 21.49 38.66 48.06 Sbj 392
15.593 7.064 3.27 12.62 13.84 17.04 30.46 Sbj 393
18.678 8.072 6.60 13.58 18.59 26.80 29.83 Sbj 394
20.621 13.560 5.58 12.21 17.50 22.40 55.41 Sbj 395
24.389 13.506 10.07 13.66 21.78 32.26 52.82 Sbj 396
------------------------------------------------------------
23.617 13.795 9.28 14.83 20.06 29.66 53.53
Chi2 for saturated model: summed across both Ss and blocks:
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
23.617 17.476 2.46 14.22 20.18 27.88 147.43
Number of significant chi2 in each block:
[12 12 12 9 10 12 6 9 0 5 4]
Number of significant chi2 in each subject:
[4 3 3 3 2 2 6 4 5 3 2 5 5 3 0 7 2 4 4 2 3 5 5 1 3 2 3]
Total number of significant chi2: 91 out of 297
acceptable_chi2 =
49.9916
Number of unacceptable chi2 in each block:
[5 0 1 3 1 1 0 2 0 0 0]
Number of unacceptable chi2 in each subject:
[0 0 0 0 1 0 2 0 3 0 0 0 0 0 0 0 0 1 0 0 3 1 0 0 0 1 1]
Total number of unacceptable chi2: 13 out of 297
The ten most deviating values:
[147.431 141.713 135.674 91.887 86.317 81.652 77.808 72.7 70.716 65.188]
G-squared statistic per subject for each of the 4 models:
Added by Alex, 2010-10-26. Updated by Alex 2010-11-06.
Per Roger's email of 2010-10-27, the Gsq value in files par11a.txt (Model 2), par11tera.txt (Model 3), and par11tersta.txt (Model 4) must be multiplied by 2. (This constant was omitted in the code that computed them on 2010-10-09.) Model 1 (which was re-fitted block by block on 2010-10-21) has the correct values because the software bug was fixed by then. 2010-11-06: The multiplication of par11tera and par11tersta is performed above -- see the sections that compare with par11terb and par11terstb.
Gsq_1 = sum(Gsq_1_per_period,2) ; % does NOT need multiplication by 2 Gsq_2 = df_2(:,end) .*2 ; % multiplied by 2 per Roger's email of 2010-10-27 Gsq_3 = df_3(:,end) ; % no need to multiply by 2, 2010-11-06 Gsq_4 = df_4(:,end) ; % no need to multiply by 2, 2010-11-06 Gsquared = [Gsq_1 Gsq_2 Gsq_3 Gsq_4 df_2(:,1)] ; % [M1 M2 M3 M4, sbj] fprintf(' Model1 Model2 Model3 Model4 G-squared \n') ; fprintf('%7.2f %7.2f %7.2f %7.2f for subject %3d \n',Gsquared') ; describe(Gsquared(:,1:4),{'Model 1', 'Model 2', 'Model 3', 'Model 4'}) % Sanity check fprintf('\n\nBecause the models are nested, the G^2 should decrease \n') ; fprintf('monotonically as the number of parameter increases. That is:\n'); fprintf('Model 1 < Model 4 < Model 3 < Model 2 \n\n') ; for k1 = 1:4 for k2 = (k1+1):4 fprintf('Model %d has lower G^2 than Model %d for %2d of the 27 Ss\n',... k1, k2, sum(Gsquared(:,k1)<Gsquared(:,k2))) ; end end clear k1 k2 % The constraint Gsquare(4) >= Gsquare(1) is violated for sbj 363: Gsquared((Gsquared(:,4)<Gsquared(:,1)),5)'
Model1 Model2 Model3 Model4 G-squared
267.52 1251.95 365.51 333.81 for subject 353
205.75 674.50 439.97 362.89 for subject 354
234.58 708.02 393.00 317.91 for subject 355
214.65 587.93 357.01 283.25 for subject 356
281.93 752.88 510.19 332.74 for subject 357
211.17 605.06 429.86 309.40 for subject 359
447.97 2476.23 1300.34 824.25 for subject 360
244.88 1072.65 538.19 331.41 for subject 362
375.69 583.35 375.50 287.25 for subject 363
238.80 1553.93 653.11 528.94 for subject 364
197.49 552.67 386.62 262.71 for subject 366
235.53 1874.95 608.50 468.22 for subject 367
270.77 1131.36 618.82 531.43 for subject 382
208.96 871.59 506.67 398.26 for subject 383
177.70 1332.98 590.98 401.22 for subject 384
303.10 941.01 536.42 420.13 for subject 385
206.72 635.38 415.43 333.06 for subject 386
355.31 2660.74 497.38 356.15 for subject 387
237.42 1169.28 409.95 357.82 for subject 388
214.96 1461.69 465.37 405.26 for subject 389
340.79 1347.14 654.44 519.12 for subject 390
358.87 1436.60 1247.09 825.78 for subject 391
311.62 1454.07 1106.72 829.73 for subject 392
171.52 1025.46 574.66 373.90 for subject 393
205.45 497.30 303.45 218.96 for subject 394
226.83 1706.92 498.83 319.19 for subject 395
268.28 1599.72 433.07 373.37 for subject 396
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
259.788 67.715 171.52 209.52 237.42 297.81 447.97 Model 1
1183.901 563.982 497.30 682.88 1131.36 1459.78 2660.74 Model 2
563.595 255.253 303.45 411.32 498.83 604.12 1300.34 Model 3
418.747 165.674 218.96 322.25 362.89 456.20 829.73 Model 4
------------------------------------------------------------
606.508 263.156 297.81 406.49 557.63 704.48 1309.69
Because the models are nested, the G^2 should decrease
monotonically as the number of parameter increases. That is:
Model 1 < Model 4 < Model 3 < Model 2
Model 1 has lower G^2 than Model 2 for 27 of the 27 Ss
Model 1 has lower G^2 than Model 3 for 26 of the 27 Ss
Model 1 has lower G^2 than Model 4 for 26 of the 27 Ss
Model 2 has lower G^2 than Model 3 for 0 of the 27 Ss
Model 2 has lower G^2 than Model 4 for 0 of the 27 Ss
Model 3 has lower G^2 than Model 4 for 0 of the 27 Ss
ans =
363
Fix the anomalous G-square values for sbj 363, MODEL 1
As of 2010-11-06, Sbj 363 violates the constraint Gsquare(4) >= Gsquare(1). There can be an unknown number of undetected errors, which we didn't detect because they didn't get so large as to reverse the inequalities.
As a temporary fix, we're just going to replace the anomalously large G-squares with those for the next model up for the same subject.
%Gsquared(:,3) = min(Gsquared(:,3),Gsquared(:,2)-.0001) ; % enforce M3 < M2 %Gsquared(:,4) = min(Gsquared(:,4),Gsquared(:,3)-.0001) ; % enforce M4 < M3 Gsquared(:,1) = min(Gsquared(:,1),Gsquared(:,4)-.0001) ; % enforce M1 < M4 % Now Model 1 should have the lowest Gsquare for each subject: assert(all(argmin(Gsquared(:,1:4)')'==1)) ; % % Print the corrected values % fprintf(' Model1 Model2 Model3 Model4 CORRECTED G-squared \n') ; % fprintf('%7.2f %7.2f %7.2f %7.2f for subject %3d \n',Gsquared') ; describe(Gsquared(:,1:4),{'Model 1', 'Model 2', 'Model 3', 'Model 4'}) % Sanity check -- this time everything should be either 0 or 27 fprintf('\n\nBecause the models are nested, the G^2 should decrease \n') ; fprintf('monotonically as the number of parameter increases. That is:\n'); fprintf('Model 1 < Model 4 < Model 3 < Model 2 \n\n') ; for k1 = 1:4 for k2 = (k1+1):4 fprintf('Model %d has lower G^2 than Model %d for %2d of the 27 Ss\n',... k1, k2, sum(Gsquared(:,k1)<Gsquared(:,k2))) ; end end clear k1 k2
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
256.512 63.926 171.52 209.52 237.42 285.92 447.97 Model 1
1183.901 563.982 497.30 682.88 1131.36 1459.78 2660.74 Model 2
563.595 255.253 303.45 411.32 498.83 604.12 1300.34 Model 3
418.747 165.674 218.96 322.25 362.89 456.20 829.73 Model 4
------------------------------------------------------------
605.689 262.209 297.81 406.49 557.63 701.51 1309.69
Because the models are nested, the G^2 should decrease
monotonically as the number of parameter increases. That is:
Model 1 < Model 4 < Model 3 < Model 2
Model 1 has lower G^2 than Model 2 for 27 of the 27 Ss
Model 1 has lower G^2 than Model 3 for 27 of the 27 Ss
Model 1 has lower G^2 than Model 4 for 27 of the 27 Ss
Model 2 has lower G^2 than Model 3 for 0 of the 27 Ss
Model 2 has lower G^2 than Model 4 for 0 of the 27 Ss
Model 3 has lower G^2 than Model 4 for 0 of the 27 Ss
Bayesian Information Criterion (BIC)
Added by Alex, 2010-10-27 See Roger & Smith (2004, Psych Review, vol 111, equations on p. 342-343)
% Fix the N term in these formulas: N = number of obesrvations per condition. % In our case N=240 because there are 480 trials per "period" (= 4 blocks) % and they are divided into 240 easy and 240 difficult trials. The % distinction between left and right stimuli is ignored in the calculation % of quantiles. Also, the "rogue" period 9 is assumed (wrongly) to have % the same N as the other 10 periods. This should not affect the outcome % because it's just 1 period out of 11, the log-likelihood part is % calculated correctly, and the model-complexity part takes log(N). N_observations_per_condition = trials_discrim_block * 4 / 2 % 240 log_N_observ = log(N_observations_per_condition) % 5.48 = log(240) BIC = NaN(size(Gsquared)) ; BIC(:,5) = Gsquared(:,5) ; % subject number % Number of parameters for each of the 4 models: % MODEL 1: Saturated model -- 7 parameters for each of the 11 blocks: % The 7 params are: v1, v2, Ter, st, sz, a, eta N_params.model1 = 7*N_periods_total ; BIC(:,1) = Gsquared(:,1) + log_N_observ*N_params.model1 ; % MODEL 2: This is the most constrained model, with the fewest params. % In MODEL_2, only drift is allowed to vary across periods. The same % boundary separation a and the same Ter and st apply across the board % for each individual subject. % This makes for 5 + 2*11 = 27 parameters N_params.model2 = 5 + 2*N_periods_total ; % 2=[v1 v2] BIC(:,2) = Gsquared(:,2) + log_N_observ*N_params.model2 ; % MODEL 3: Both drift and Ter are allowed to vary across periods. % The same boundary separation a and the same st apply across the board % for each individual subject. % This makes for 4 + 3*11 = 37 parameters N_params.model3 = 4 + 3*N_periods_total ; % 3=[v1 v2 Ter] BIC(:,3) = Gsquared(:,3) + log_N_observ*N_params.model3 ; % MODEL_4: Three things (4 parameters total because there is easy and % hard drift) are allowed to vary across periods. These are: % - drift rate (easy and hard) % - mean nondecision time Ter % - range of the nondecision time distribution st % The same boundary separation a applies across the board for each % individual subject. Also the same eta and sz. % This makes for 3 + 4*11 = 47 parameters N_params.model4 = 3 + 4*N_periods_total % 4=[v1 v2 Ter st] BIC(:,4) = Gsquared(:,4) + log_N_observ*N_params.model4 ; % The "winner" model for each subject is the one with the lowest BIC: BIC_winner = argmin(BIC(:,1:4)')' ; fprintf('**** Approximate BIC *****\n') ; fprintf(' Model1 Model2 Model3 Model4 =Sbj= Win\n') ; fprintf('%7.2f %7.2f %7.2f %7.2f =%3d= %d \n',[BIC BIC_winner]') ; fprintf(' Model1 Model2 Model3 Model4 =Sbj= Win\n') ; describe(BIC(:,1:4),{'Model 1', 'Model 2', 'Model 3', 'Model 4'}) xtab1(BIC_winner)
N_observations_per_condition =
240
log_N_observ =
5.4806
N_params =
model1: 77
model2: 27
model3: 37
model4: 47
**** Approximate BIC *****
Model1 Model2 Model3 Model4 =Sbj= Win
689.53 1399.92 568.30 591.40 =353= 3
627.76 822.47 642.76 620.48 =354= 4
656.59 856.00 595.78 575.50 =355= 4
636.66 735.91 559.80 540.84 =356= 4
703.94 900.85 712.97 590.33 =357= 4
633.18 753.04 632.64 566.99 =359= 4
869.98 2624.20 1503.13 1081.85 =360= 1
666.89 1220.63 740.98 589.00 =362= 4
709.26 731.33 578.29 544.84 =363= 4
660.81 1701.91 855.89 786.53 =364= 1
619.50 700.65 589.40 520.30 =366= 4
657.54 2022.93 811.28 725.81 =367= 1
692.78 1279.34 821.61 789.03 =382= 1
630.97 1019.57 709.45 655.85 =383= 1
599.71 1480.96 793.76 658.81 =384= 1
725.11 1088.99 739.21 677.72 =385= 4
628.73 783.36 618.21 590.65 =386= 4
777.32 2808.72 700.16 613.74 =387= 4
659.43 1317.26 612.73 615.41 =388= 3
636.96 1609.67 668.15 662.85 =389= 1
762.80 1495.12 857.22 776.71 =390= 1
780.88 1584.57 1449.87 1083.37 =391= 1
733.63 1602.05 1309.50 1087.32 =392= 1
593.53 1173.44 777.44 631.49 =393= 1
627.46 645.28 506.23 476.55 =394= 4
648.84 1854.90 701.62 576.78 =395= 4
690.29 1747.70 635.86 630.96 =396= 4
Model1 Model2 Model3 Model4 =Sbj= Win
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
678.521 63.926 593.53 631.52 659.43 707.93 869.98 Model 1
1331.879 563.982 645.28 830.85 1279.34 1607.76 2808.72 Model 2
766.379 255.253 506.23 614.10 701.62 806.90 1503.13 Model 3
676.337 165.674 476.55 579.84 620.48 713.79 1087.32 Model 4
------------------------------------------------------------
863.279 262.209 555.40 664.08 815.22 959.10 1567.28
Value Count Percent Cum_cnt Cum_pct
-------------------------------------------
1 11 40.74 11 40.74
3 2 7.41 13 48.15
4 14 51.85 27 100.00
-------------------------------------------
BIC competition outcomes counted across the 27 subjects
Added by Alex, 2010-10-27 For BIC, lower is better.
for k1 = 1:4 for k2 = (k1+1):4 fprintf('Model %d has lower BIC than Model %d for %2d of the 27 Ss\n',... k1, k2, sum(BIC(:,k1)<BIC(:,k2))) ; end end clear k1 k2
Model 1 has lower BIC than Model 2 for 27 of the 27 Ss Model 1 has lower BIC than Model 3 for 16 of the 27 Ss Model 1 has lower BIC than Model 4 for 11 of the 27 Ss Model 2 has lower BIC than Model 3 for 0 of the 27 Ss Model 2 has lower BIC than Model 4 for 0 of the 27 Ss Model 3 has lower BIC than Model 4 for 2 of the 27 Ss
Specificity Indices (for all sbjs averaged together)
plus confidence interval calculations This section is old. Was updated to 11 periods 2010-10-20.
% Grab all sbjs' dprime data by block adpr = [H.all_dprime] ; % [40, N_subjects] edpr = [H.easy_dprime] ; hdpr = [H.hard_dprime] ; adpr_mean = mean(adpr,2) ; edpr_mean = mean(edpr,2) ; hdpr_mean = mean(hdpr,2) ; % Aggregate blocks into 11 "periods": [40, N_sbj] --> [11, N_sbj] dp11 = [mean(adpr(1:4,:),1);... % Session 1 (1/2) mean(adpr(5:8,:),1);... % Session 1 (2/2) mean(adpr(9:12,:),1);... % Session 2 (1/2) ... mean(adpr(13:16,:),1);... % Session 2 (2/2) mean(adpr(17:20,:),1);... % Session 3 (1/2) ... mean(adpr(21:24,:),1);... % Session 3 (2/2) mean(adpr(25:28,:),1);... % Session 4 (1/2) ... mean(adpr(29:32,:),1);... % Session 4 (2/2) mean(adpr(33,:),1);... % Session 5 *block 1* (1/3) mean(adpr(34:37,:),1);... % Session 5 (2/3) mean(adpr(38:40,:),1)]; % Session 5 (3/3) edp11 = [mean(edpr(1:4,:),1);... % Session 1 (1/2) mean(edpr(5:8,:),1);... % Session 1 (2/2) mean(edpr(9:12,:),1);... % Session 2 (1/2) ... mean(edpr(13:16,:),1);... % Session 2 (2/2) mean(edpr(17:20,:),1);... % Session 3 (1/2) ... mean(edpr(21:24,:),1);... % Session 3 (2/2) mean(edpr(25:28,:),1);... % Session 4 (1/2) ... mean(edpr(29:32,:),1);... % Session 4 (2/2) mean(edpr(33,:),1);... % Session 5 *block 1* (1/3) mean(edpr(34:37,:),1);... % Session 5 (2/3) mean(edpr(38:40,:),1)]; % Session 5 (3/3) hdp11 = [mean(hdpr(1:4,:),1);... % Session 1 (1/2) mean(hdpr(5:8,:),1);... % Session 1 (2/2) mean(hdpr(9:12,:),1);... % Session 2 (1/2) ... mean(hdpr(13:16,:),1);... % Session 2 (2/2) mean(hdpr(17:20,:),1);... % Session 3 (1/2) ... mean(hdpr(21:24,:),1);... % Session 3 (2/2) mean(hdpr(25:28,:),1);... % Session 4 (1/2) ... mean(hdpr(29:32,:),1);... % Session 4 (2/2) mean(hdpr(33,:),1);... % Session 5 *block 1* (1/3) mean(hdpr(34:37,:),1);... % Session 5 (2/3) mean(hdpr(38:40,:),1)]; % Session 5 (3/3) % Avg RT across all subjects aRT_all = mean(sRT_all,1) ; aRT_corr = mean(sRT_corr,1) ; aRTsd_all = mean(RTsd_all,1) ; aRTsd_corr = mean(RTsd_corr,1) ;
dprime confidence interval calculations
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% % This section is old. Was updated to 11 periods 2010-10-20. % Subtract individual subjects' means to estimate within-sbj variance % All dprime madpr_by_sbj = mean(dp11); % [1 x N_subjects] adpr_centered = dp11 - repmat(madpr_by_sbj,N_periods_total,1) ; semadpr = std(adpr_centered,0,2); % Easy dprime medpr_by_sbj = mean(edp11); % [1 x N_subjects] edpr_centered = edp11 - repmat(medpr_by_sbj,N_periods_total,1) ; semedpr = std(edpr_centered,0,2); % Hard dprime mhdpr_by_sbj = mean(hdp11); % [1 x N_subjects] hdpr_centered = hdp11 - repmat(mhdpr_by_sbj,N_periods_total,1) ; semhdpr = std(hdpr_centered,0,2); % within-sbj confidence intervals alpha = .90 ; % confidence level z_crit = norminv((1+alpha)/2) ; % 1.64 for alpha=90% CI_madpr = z_crit.*(semadpr./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_medpr = z_crit.*(semedpr./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_mhdpr = z_crit.*(semhdpr./repmat(sqrt(N_sbj),N_periods_total,1)) ;
RT confidence interval calculations
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% % This section is old. Was updated to 11 periods 2010-10-20. % All RTs RT_all_centered = sRT_all - repmat(aRT_all, 27, 1); seRT_all = std(RT_all_centered,0,1) ; % Correct RTs RT_corr_centered = sRT_corr - repmat(aRT_corr, 27, 1); seRT_corr = std(RT_corr_centered,0,1) ; % All RT std RTsd_all_centered = RTsd_all - repmat(aRTsd_all, 27, 1); seRTsd_all = std(RTsd_all_centered,0,1) ; % Correct RT std RTsd_corr_centered = RTsd_corr - repmat(aRTsd_corr, 27, 1); seRTsd_corr = std(RTsd_corr_centered,0,1) ; % within-sbj confidence intervals CI_RT_all = z_crit.*(seRT_all./repmat(sqrt(N_sbj),1,N_periods_total)) ; CI_RT_corr = z_crit.*(seRT_corr./repmat(sqrt(N_sbj),1,N_periods_total)) ; CI_RTsd_all = z_crit.*(seRTsd_all./repmat(sqrt(N_sbj),1,N_periods_total)) ; CI_RTsd_corr = z_crit.*(seRTsd_corr./repmat(sqrt(N_sbj),1,N_periods_total)) ;
diffusion confidence interval calculations
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% % This section is old. Was updated to 11 periods 2010-10-20. % This is about MODEL_1 -- the saturated model. The other diffusion models % do not have enough parameters to calculate confidence intervals. % MODELs 2 through 4 get their confidence intervals through bootstrapping. % The current section was written before there was a bootstrap. % Subtract individual subjects' means to estimate within-sbj variance % a ma_by_sbj = mean(sbj_means(:,a),2)' ; % [N_periods_total x N_subjects] a_centered = sbj_means(:,a)' - repmat(ma_by_sbj, N_periods_total, 1) ; sea = std(a_centered,0,2) ; % ter mter_by_sbj = mean(sbj_means(:,ter),2)' ; % [N_periods_total x N_subjects] ter_centered = sbj_means(:,ter)' - repmat(mter_by_sbj, N_periods_total, 1) ; seter = std(ter_centered,0,2) ; % eta meta_by_sbj = mean(sbj_means(:,eta),2)' ; % [N_periods_total x N_subjects] eta_centered = sbj_means(:,eta)' - repmat(meta_by_sbj, N_periods_total, 1) ; seeta = std(eta_centered,0,2) ; % sz msz_by_sbj = mean(sbj_means(:,sz),2)' ; % [N_periods_total x N_subjects] sz_centered = sbj_means(:,sz)' - repmat(msz_by_sbj, N_periods_total, 1) ; sesz = std(sz_centered,0,2) ; % st mst_by_sbj = mean(sbj_means(:,st),2)' ; % [N_periods_total x N_subjects] st_centered = sbj_means(:,st)' - repmat(mst_by_sbj, N_periods_total, 1) ; sest = std(st_centered,0,2) ; % v1 mv1_by_sbj = mean(sbj_means(:,v1),2)' ; % [N_periods_total x N_subjects] v1_centered = sbj_means(:,v1)' - repmat(mv1_by_sbj, N_periods_total, 1) ; sev1 = std(v1_centered,0,2) ; % v2 mv2_by_sbj = mean(sbj_means(:,v2),2)' ; % [N_periods_total x N_subjects] v2_centered = sbj_means(:,v2)' - repmat(mv2_by_sbj, N_periods_total, 1) ; sev2 = std(v2_centered,0,2) ; % Tmin = t1 = Ter-st/2 [Added by Alex 2010-11-17] Tmin = sbj_means(:,ter)' - sbj_means(:,st)'./2 ; % [N_periods_total x N_subjects] mTmin_by_sbj = mean(Tmin) ; Tmin_centered = Tmin - repmat(mTmin_by_sbj, N_periods_total, 1) ; seTmin = std(Tmin_centered,0,2) ; % within-sbj confidence intervals CI_a = z_crit.*(sea./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_ter = z_crit.*(seter./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_eta = z_crit.*(seeta./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_sz = z_crit.*(sesz./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_st = z_crit.*(sest./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_v1 = z_crit.*(sev1./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_v2 = z_crit.*(sev2./repmat(sqrt(N_sbj),N_periods_total,1)) ; CI_Tmin = z_crit.*(seTmin./repmat(sqrt(N_sbj),N_periods_total,1)) ; % Average over subject dprimes (for SI calculations and plotting purposes) mdp11 = mean(dp11,2) ; medp11 = mean(edp11,2) ; mhdp11 = mean(hdp11,2) ; % RT Specificity Index (SI) % SI_RT_all = (aRT_all(5) - aRT_all(7)) / ... % (aRT_all(5) - aRT_all(1)); % SI_RT_corr = (aRT_corr(5) - aRT_corr(7)) / ... % (aRT_corr(5) - aRT_corr(1)); SI_RT_all = specif_idx(aRT_all); SI_RT_corr = specif_idx(aRT_corr); % RT std Specificity Index (SI) SI_RTsd_all = specif_idx(aRTsd_all); SI_RTsd_corr = specif_idx(aRTsd_corr); % Dprime Specificity Index (SI) SI_dpr = specif_idx(mdp11'); % Easy Dprime Specificity Index (SI) SI_edpr = specif_idx(medp11'); % Hard Dprime Specificity Index (SI) SI_hdpr = specif_idx(mhdp11'); % Parameter a Specificity Index SI_a = specif_idx(mean(sbj_means(:, a))); % Parameter ter Specificity Index SI_ter = specif_idx(mean(sbj_means(:, ter))); % Parameter eta Specificity Index SI_eta = specif_idx(mean(sbj_means(:, eta))); % Parameter sz Specificity Index SI_sz = specif_idx(mean(sbj_means(:, sz))); % Parameter st Specificity Index SI_st = specif_idx(mean(sbj_means(:, st))); % Drift rate 1 Specificity Index SI_v1 = specif_idx(mean(sbj_means(:, v1))); % Drift rate 2 Specificity Index SI_v2 = specif_idx(mean(sbj_means(:, v2)));
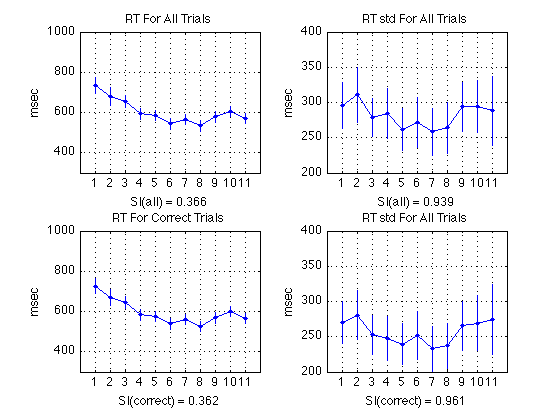
Plot RT and RT std (sbjs averaged together)
% All RTs subplot(2,2,1) ; plot(1:N_periods_total, aRT_all, 'b.-') ; axis([0 N_periods_total+1 300 1000]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT For All Trials') ; ylabel('msec') ; xlabel(sprintf('SI(all) = %0.3f', SI_RT_all)); errorbar1(1:N_periods_total,aRT_all,CI_RT_all,['b' 'n']) ; % Correct RTs subplot(2,2,3) ; plot(1:N_periods_total, aRT_corr, 'b.-') ; axis([0 N_periods_total+1 300 1000]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT For Correct Trials') ; ylabel('msec') ; xlabel(sprintf('SI(correct) = %0.3f', SI_RT_corr)); errorbar1(1:N_periods_total,aRT_corr,CI_RT_corr,['b' 'n']) ; % All RT std subplot(2,2,2) ; plot(1:N_periods_total, aRTsd_all, 'b.-') ; axis([0 N_periods_total+1 200 400]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT std For All Trials') ; ylabel('msec') ; xlabel(sprintf('SI(all) = %0.3f', SI_RTsd_all)); errorbar1(1:N_periods_total,aRTsd_all,CI_RTsd_all,['b' 'n']) ; % Correct RT std subplot(2,2,4) ; plot(1:N_periods_total, aRTsd_corr, 'b.-') ; axis([0 N_periods_total+1 200 400]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT std For All Trials') ; ylabel('msec') ; xlabel(sprintf('SI(correct) = %0.3f', SI_RTsd_corr)); errorbar1(1:N_periods_total,aRTsd_corr,CI_RTsd_corr,['b' 'n']) ;
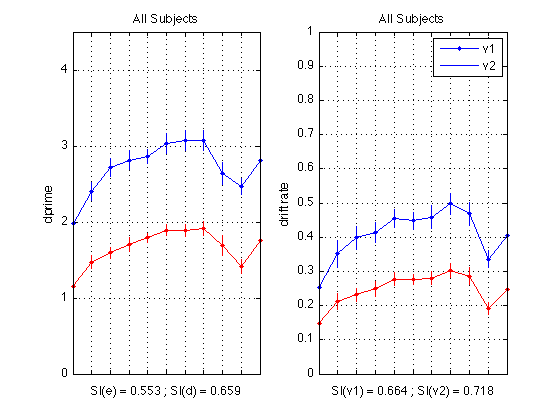
Plot dprime
CI = CI_madpr ; subplot(1,2,1) ; plot(1:N_periods_total, mdp11,'b.-') ; axis([1 N_periods_total 0 M]) ; grid on ; set(gca,'xtick',([1:N_periods_total]),... 'xticklabel',[],'ytick',(0:ceil(M))) ; title('All Subjects') ; ylabel('dprime'); xlabel(sprintf('SI = %0.3f', SI_dpr)); errorbar1(1:N_periods_total,mdp11,CI,['b' 'n']) ; % Plot drift rates subplot(1,2,2) ; plot(1:N_periods_total, mean(sbj_means(:,v1)), 'b.-'); CI = CI_v1 ; errorbar1(1:N_periods_total, mean(sbj_means(:,v1)), CI, ['b' 'n']); hold on plot(1:N_periods_total, mean(sbj_means(:,v2)), 'r.-'); CI = CI_v2 ; errorbar1(1:N_periods_total, mean(sbj_means(:,v2)), CI, ['r' 'n']); axis([1 N_periods_total 0 1]) ; grid on ; set(gca,'xtick',([1:N_periods_total]),... 'xticklabel',[],'ytick',(0:.1:1)) ; title('All Subjects') ; ylabel('drift rate'); xlabel(sprintf('SI(v1) = %0.3f ; SI(v2) = %0.3f', SI_v1, SI_v2)); legend('v1','v2','location','NorthEast'); hold off
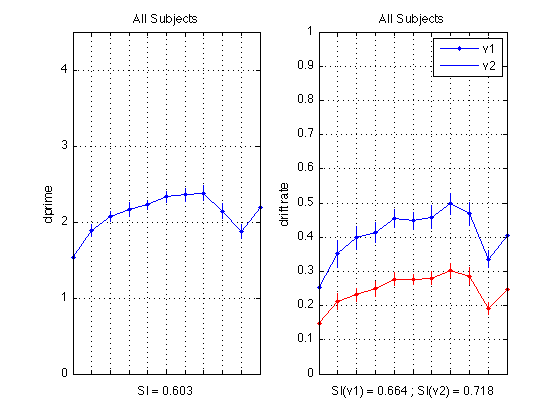
All subjects plot with separate deltas
Delta is easy=7 degrees versus difficult=4 degrees difference b/n clockwise and counterclockwise stimuli.
% Plot dprime subplot(1,2,1) ; plot(1:N_periods_total, medp11,'b.-') ; CI = CI_medpr ; errorbar1(1:N_periods_total,medp11,CI,['b' 'n']) ; hold on plot(1:N_periods_total, mhdp11,'r.-') ; CI = CI_mhdpr ; errorbar1(1:N_periods_total,mhdp11,CI,['r' 'n']) ; axis([1 N_periods_total 0 M]) ; grid on ; set(gca,'xtick',([1:N_periods_total]),... 'xticklabel',[],'ytick',(0:ceil(M))) ; title('All Subjects') ; ylabel('dprime'); xlabel(sprintf('SI(e) = %0.3f ; SI(d) = %0.3f', SI_edpr, SI_hdpr)); % Plot drift rates subplot(1,2,2) ; plot(1:N_periods_total, mean(sbj_means(:,v1)), 'b.-'); CI = CI_v1 ; errorbar1(1:N_periods_total, mean(sbj_means(:,v1)), CI, ['b' 'n']); hold on plot(1:N_periods_total, mean(sbj_means(:,v2)), 'r.-'); CI = CI_v2 ; errorbar1(1:N_periods_total, mean(sbj_means(:,v2)), CI, ['r' 'n']); axis([1 N_periods_total 0 1]) ; grid on ; set(gca,'xtick',([1:N_periods_total]),... 'xticklabel',[],'ytick',(0:.1:1)) ; title('All Subjects') ; ylabel('drift rate'); xlabel(sprintf('SI(v1) = %0.3f ; SI(v2) = %0.3f', SI_v1, SI_v2)); legend('v1','v2','location','NorthEast'); hold off
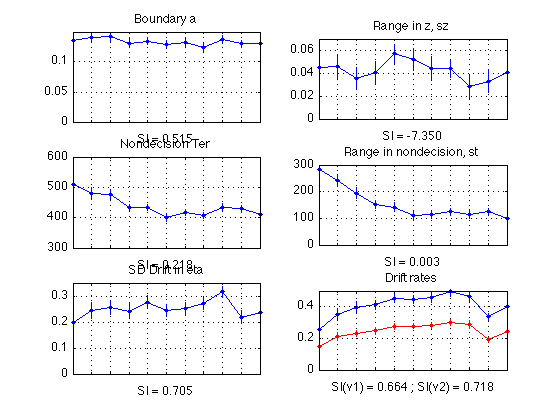
Plot diffusion params (averaged across all sbjs)
MODEL_1, non-bootstrapped, updated for 11 periods, as of 2010-10-21
subplot(3,2,1) ; % a plot(1:N_periods_total, mean(sbj_means(:, a)), 'b.-'); CI = CI_a ; errorbar1(1:N_periods_total, mean(sbj_means(:,a)), CI, ['b' 'n']); title('Boundary a'); xlabel(sprintf('SI = %0.3f', mean(SI_a))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 0.15]) ; grid on ; subplot(3,2,3) ; % ter plot(1:N_periods_total, mean(sbj_means(:, ter)*1000), 'b.-'); CI = CI_ter ; errorbar1(1:N_periods_total, mean(sbj_means(:,ter)*1000), CI*1000, ['b' 'n']); title('Nondecision Ter'); xlabel(sprintf('SI = %0.3f', mean(SI_ter))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 300 600]) ; grid on ; subplot(3,2,5) ; % eta plot(1:N_periods_total, mean(sbj_means(:, eta)), 'b.-'); CI = CI_eta ; errorbar1(1:N_periods_total, mean(sbj_means(:,eta)), CI, ['b' 'n']); title('SD Drift in eta'); xlabel(sprintf('SI = %0.3f', mean(SI_eta))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 0.35]) ; grid on ; subplot(3,2,2) ; % sz plot(1:N_periods_total, mean(sbj_means(:, sz)), 'b.-'); CI = CI_sz ; errorbar1(1:N_periods_total, mean(sbj_means(:,sz)), CI, ['b' 'n']); title('Range in z, sz'); xlabel(sprintf('SI = %0.3f', mean(SI_sz))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 0.07]) ; grid on ; subplot(3,2,4) ; % st plot(1:N_periods_total, mean(sbj_means(:, st)*1000), 'b.-'); CI = CI_st ; errorbar1(1:N_periods_total, mean(sbj_means(:,st)*1000), CI*1000, ['b' 'n']); title('Range in nondecision, st'); xlabel(sprintf('SI = %0.3f', mean(SI_st))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 300]) ; grid on ; % v1 subplot(3,2,6) ; plot(1:N_periods_total, mean(sbj_means(:, v1)), 'b.-'); CI = CI_v1 ; errorbar1(1:N_periods_total, mean(sbj_means(:,v1)), CI, ['b' 'n']); title('Drift rates'); xlabel(sprintf('SI(v1) = %0.3f ; SI(v2) = %0.3f', mean(SI_v1),mean(SI_v2))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); hold on % v2 plot(1:N_periods_total, mean(sbj_means(:, v2)), 'r.-'); CI = CI_v2 ; errorbar1(1:N_periods_total, mean(sbj_means(:,v2)), CI, ['r' 'n']); axis([1 N_periods_total 0 0.5]) ; grid on ; hold off
Export data for the six-plot empirical figure, 2010-11-17
These data were ported to R and used to generate the main data figure for the revised manuscript sent to PB&R in November 2010. They are the group-averaged parameters for the saturated model (MODEL_1) across the 11 time periods.
% Panel A fprintf('\n\nEasy dprime: Group mean: medp11 = mean(edp11,2)\n') ; fprintf(' %6.4f',medp11) ; fprintf('\n CI90 w/in sbj: CI_medpr\n') ; fprintf(' %6.4f',CI_medpr) ; fprintf('\n\nHard dprime: Group mean: medp11 = mean(hdp11,2)\n') ; fprintf(' %6.4f',mhdp11) ; fprintf('\n CI90 w/in sbj: CI_mhdpr\n') ; fprintf(' %6.4f',CI_mhdpr) ; fprintf('\n\nAverage dprime (not plotted): Group mean: mdp11 = mean(dp11,2)\n') ; fprintf(' %6.4f',mdp11) ; fprintf('\n CI90 w/in sbj: CI_madpr\n') ; fprintf(' %6.4f',CI_madpr) ; % Panel B fprintf('\n\nAverage mean RT (all trials): Group mean: aRT_all = mean(sRT_all,1)\n') ; fprintf(' %6.1f',aRT_all) ; fprintf('\n CI90 w/in sbj: CI_RT_all\n') ; fprintf(' %6.2f',CI_RT_all) ; % Panel C fprintf('\n\nEasy drift rate v1: mean(sbj_means(:, v1))\n') ; fprintf(' %6.4f',mean(sbj_means(:, v1))) ; fprintf('\n CI90 w/in sbj: CI_v1\n') ; fprintf(' %6.4f',CI_v1) ; fprintf('\n\nHard drift rate v2: mean(sbj_means(:, v2))\n') ; fprintf(' %6.4f',mean(sbj_means(:, v2))) ; fprintf('\n CI90 w/in sbj: CI_v2\n') ; fprintf(' %6.4f',CI_v2) ; % Panel D fprintf('\n\nMean nondecision time Ter: mean(sbj_means(:, ter)*1000)\n') ; fprintf(' %6.1f',mean(sbj_means(:, ter)*1000)) ; fprintf('\n CI90 w/in sbj: CI_ter*1000\n') ; fprintf(' %6.2f',CI_ter*1000) ; fprintf('\n\nRange in nondecision time st (not plotted): mean(sbj_means(:, st)*1000)\n') ; fprintf(' %6.1f',mean(sbj_means(:, st)*1000)) ; fprintf('\n CI90 w/in sbj: CI_st*1000\n') ; fprintf(' %6.2f',CI_st*1000) ; fprintf('\n\nMinimum nondecision time Tmin=t1: mean(Tmin,2)*1000\n') ; fprintf(' %6.1f',mean(Tmin,2)*1000) ; fprintf('\n CI90 w/in sbj: CI_Tmin*1000\n') ; fprintf(' %6.2f',CI_Tmin*1000) ; % Panel E fprintf('\n\nVariability in drift rate eta: mean(sbj_means(:, eta))\n') ; fprintf(' %6.4f',mean(sbj_means(:, eta))) ; fprintf('\n CI90 w/in sbj: CI_eta\n') ; fprintf(' %6.4f',CI_eta) ; % Panel F fprintf('\n\nBoundary separation a: mean(sbj_means(:, a))\n') ; fprintf(' %6.4f',mean(sbj_means(:, a))) ; fprintf('\n CI90 w/in sbj: CI_a\n') ; fprintf(' %6.4f',CI_a) ; % Not plotted fprintf('\n\nRange in starting point sz: mean(sbj_means(:, sz))\n') ; fprintf(' %6.4f',mean(sbj_means(:, sz))) ; fprintf('\n CI90 w/in sbj: CI_sz\n') ; fprintf(' %6.4f',CI_sz) ; fprintf('\n\nMAESpec_diffusion_fits executed on %s.\n\n',datestr(now)) ;
Easy dprime: Group mean: medp11 = mean(edp11,2) 1.9742 2.4024 2.7152 2.8128 2.8620 3.0312 3.0682 3.0706 2.6388 2.4638 2.8112 CI90 w/in sbj: CI_medpr 0.0946 0.1233 0.1219 0.1256 0.0949 0.1272 0.1314 0.1287 0.1476 0.1076 0.1409 Hard dprime: Group mean: medp11 = mean(hdp11,2) 1.1553 1.4729 1.5972 1.7069 1.7923 1.8890 1.8887 1.9091 1.6944 1.4125 1.7599 CI90 w/in sbj: CI_mhdpr 0.0773 0.0821 0.0729 0.0946 0.0602 0.0731 0.0846 0.0912 0.1305 0.0957 0.0868 Average dprime (not plotted): Group mean: mdp11 = mean(dp11,2) 1.5379 1.8925 2.0733 2.1644 2.2251 2.3292 2.3588 2.3805 2.1424 1.8721 2.1925 CI90 w/in sbj: CI_madpr 0.0746 0.0791 0.0674 0.0969 0.0577 0.0688 0.0854 0.0966 0.1029 0.0914 0.0901 Average mean RT (all trials): Group mean: aRT_all = mean(sRT_all,1) 734.2 679.7 655.1 594.0 583.7 543.3 566.7 533.5 579.3 607.0 571.9 CI90 w/in sbj: CI_RT_all 39.64 42.95 29.79 28.62 24.68 25.85 25.90 26.11 26.81 25.60 24.72 Easy drift rate v1: mean(sbj_means(:, v1)) 0.2541 0.3517 0.3980 0.4133 0.4546 0.4477 0.4588 0.4973 0.4682 0.3358 0.4050 CI90 w/in sbj: CI_v1 0.0280 0.0391 0.0337 0.0297 0.0267 0.0252 0.0336 0.0305 0.0332 0.0244 0.0312 Hard drift rate v2: mean(sbj_means(:, v2)) 0.1477 0.2118 0.2311 0.2497 0.2776 0.2769 0.2798 0.3023 0.2855 0.1913 0.2461 CI90 w/in sbj: CI_v2 0.0184 0.0243 0.0199 0.0221 0.0192 0.0145 0.0188 0.0204 0.0251 0.0166 0.0191 Mean nondecision time Ter: mean(sbj_means(:, ter)*1000) 510.0 479.9 477.6 433.8 433.4 400.6 416.4 406.0 433.3 428.6 409.0 CI90 w/in sbj: CI_ter*1000 26.19 21.09 16.37 9.19 8.98 12.81 10.30 9.44 12.85 10.84 13.18 Range in nondecision time st (not plotted): mean(sbj_means(:, st)*1000) 282.7 241.6 194.6 151.1 141.6 110.7 115.8 124.9 115.6 125.4 99.6 CI90 w/in sbj: CI_st*1000 28.45 22.91 21.61 15.39 17.78 13.87 12.67 13.26 16.76 11.55 16.41 Minimum nondecision time Tmin=t1: mean(Tmin,2)*1000 368.6 359.1 380.3 358.2 362.6 345.3 358.5 343.5 375.5 365.9 359.2 CI90 w/in sbj: CI_Tmin*1000 35.95 19.85 17.48 8.03 8.92 12.27 10.11 9.68 16.22 10.18 13.63 Variability in drift rate eta: mean(sbj_means(:, eta)) 0.1974 0.2446 0.2556 0.2412 0.2765 0.2433 0.2500 0.2707 0.3159 0.2191 0.2370 CI90 w/in sbj: CI_eta 0.0314 0.0298 0.0265 0.0295 0.0251 0.0240 0.0254 0.0226 0.0214 0.0268 0.0303 Boundary separation a: mean(sbj_means(:, a)) 0.1361 0.1415 0.1418 0.1316 0.1346 0.1292 0.1331 0.1238 0.1380 0.1301 0.1311 CI90 w/in sbj: CI_a 0.0099 0.0084 0.0068 0.0069 0.0056 0.0067 0.0069 0.0064 0.0065 0.0053 0.0062 Range in starting point sz: mean(sbj_means(:, sz)) 0.0454 0.0458 0.0353 0.0407 0.0570 0.0521 0.0440 0.0440 0.0284 0.0331 0.0411 CI90 w/in sbj: CI_sz 0.0105 0.0106 0.0097 0.0094 0.0084 0.0092 0.0106 0.0074 0.0104 0.0102 0.0091 MAESpec_diffusion_fits executed on 29-Nov-2010 17:38:51.
Raw specificity values
'Raw' in the sense that no bootstrapping is involved. Each row printed below represents a subject. Columns are: 1 = dprime 2 = a 3 = Ter 4 = eta 5 = sz 6 = st 7 = v1 8 = v2
% The following indices are calculated % As of Oct 2010, this code has not been changed since Aug 2010: % % SI: Indices with session 4 used as the final day of training and the % first half of session 5 used as the first day of transfer (minus % the first block) % SIb: Indices with session 5, block 1 used as the final day of training % and the first half of session 5 used as the first day of transfer % (minus the first block) % SIc: Indices with session 4, used as the final day of training and the % first block of session 5 used as the first day of "transfer" % (although this is not the transfer direction, but a continuation of % the training direction occurring after a session of MAE trials) SI %#ok<*NOPTS> SIb SIc
SI =
-0.2607 1.3000 0.0802 1.8889 0.5181 -0.4750 0.6696 0.5161
0.6547 0.9167 0.1774 1.6522 -0.2778 0.2548 1.1331 0.9415
0.7298 -0.9167 0 1.2553 -0.1034 -0.1894 0.4228 1.2258
0.2035 0.7711 1.7727 0.9346 Inf -0.1444 0.8756 0.8333
0.4970 -0.6444 0.1138 0 0.9615 0.0767 0.4468 0.3745
0.8770 -1.2222 0.0857 2.2973 -1.2000 -0.1172 0.9319 1.1188
0.6892 0.0328 1.6500 0.6055 0 0.1038 0.6734 0.7415
-0.1177 -0.1364 0.1327 0 0 -0.0844 -0.0605 -0.0778
0.9452 -3.8000 0.9577 1.0065 -Inf 0.8957 2.0185 0.6988
0.4311 0.2297 0.1311 NaN -0.1452 0.1517 0.1897 0.4954
0.6626 -0.2500 -0.1923 Inf 1.0000 0.2038 1.0489 0.8302
1.6666 0.6714 0.2077 0.4136 -0.7778 9.5000 0.8798 1.0328
0.5775 0.3750 0.4225 4.0714 0.5098 -0.0171 0.2868 0.5665
-0.2710 0.7838 2.8000 0.5263 0.7619 -0.0776 0.2710 0.4237
1.3976 0.1667 -0.0441 0.4884 0.1800 0.0144 0.7870 0.8058
0.1808 0.3731 0.2256 0.2727 0 -0.0636 0.5357 0.3448
0.8652 0 0.1839 -0.1895 -0.0388 -0.1286 0.6509 0.6864
1.2435 0.4694 -0.0945 Inf 0.6500 0.0916 1.0652 1.0762
0.2290 0.2000 0.0412 -1.6885 1.0000 0.1333 2.4921 2.5306
-0.7018 -1.4615 0 -15.6429 4.9286 -0.7632 2.2975 3.8750
0.5956 -5.0000 0.2840 28.6667 -4.0000 0.3663 1.1471 1.2000
0.0224 0.1892 0.0940 0.3533 -1.5172 -0.3412 0.4365 0.3806
1.3661 -1.2800 0.7101 0 -0.5893 -0.0247 0.2194 0.6243
0.6933 -0.1389 0.8462 0.5948 0 0.2233 0.8877 0.8919
0.5132 1.5455 0.1111 -0.1825 -1.6667 0.1024 0.3266 0.4081
0.5948 0.5536 0.0264 1.0556 -2.7222 -0.2554 -1.4634 0.3514
2.3037 -1.9444 0.1211 -1.0063 8.8750 0.1453 -0.3471 -1.1000
SIb =
-0.5717 0.9062 -0.1814 1.1622 0.6226 -1.0345 0.8348 0.7345
0.8224 1.0159 -0.0200 1.5310 -0.0147 -0.0473 1.1277 0.9588
0.6772 -0.3529 -0.1429 -3.0000 -Inf -0.4439 0.4538 -0.4000
0.2808 0.6984 0.7792 0.9296 2.9143 0.2961 0.9051 0.7222
-0.9064 0.0263 0.0763 0 -Inf 0.2136 -0.1094 -0.2836
0.7626 -0.2903 0.4921 2.2973 0.7027 -0.2361 0.8932 1.1739
0.5443 -0.9667 0.0250 0.7471 6.0000 0.0717 0.6957 0.7551
-0.1602 -2.1250 -6.7273 0 0 0.3949 -0.0273 -0.0112
0.8829 0.7000 0.9423 1.0274 -9.3333 0.7536 1.9016 1.2907
-0.5315 -0.3902 0.1264 NaN -Inf 0.0682 0.1856 -0.8644
0.5400 3.5000 -4.1667 0.8688 1.0000 0.5098 1.5625 3.2500
-6.1749 0.2813 0.2426 0.4974 -Inf -2.0357 0.8372 1.5714
0.4931 0.1818 0.0682 0.6417 0.7396 0.0556 0.2581 0.6364
-1.5157 0.4286 -1.5714 0.7300 0.9123 0.1935 0.2427 0.0377
1.4272 0.5000 -0.1639 0.4907 -0.2424 -0.2018 0.8145 0.8050
-0.1316 0.4545 0.2536 0.0345 0 0.2812 -0.0777 -0.0857
0.3192 0.2667 0.1839 0 0 0.1124 0.2377 0.2697
2.2789 -1.1667 -0.1583 1.0283 0.8923 0.0165 1.0898 1.0597
0.1014 6.0000 -1.1136 -4.4667 NaN 0.2778 2.6207 -1.0270
-0.4198 -3.5714 0.0780 -0.3314 0 -1.7917 -0.1716 -0.5753
0.5772 -0.2414 0.3980 2.2029 2.0526 0.1351 1.0820 1.1532
0.5466 -0.8182 -0.0495 0.6151 -1.4333 -0.0560 0.6606 0.6706
1.1675 -0.1176 1.5714 0 -0.7800 -0.0473 0.4158 0.7948
0.3447 -0.2424 2.5000 0.6737 0 -0.0335 0.8834 0.8660
0.2741 0.4545 0.2000 0 1.4000 0.3294 0.0163 -0.0126
0.6248 0.3902 -0.4832 0.9747 -0.1552 -0.2419 -0.0100 0.8812
1.2691 8.5714 0.0044 Inf 3.8636 -0.2645 -1.8596 1.6364
SIc =
0.1979 4.2000 0.2214 -4.4815 -0.2771 0.2750 -1.0000 -0.8226
-0.9441 6.2500 0.1935 -0.2283 -0.2593 0.2885 -0.0418 -0.4211
0.1631 -0.4167 0.1250 1.0638 1.0000 0.1762 -0.0569 1.1613
-0.1074 0.2410 4.5000 0.0719 -Inf -0.6257 -0.3111 0.4000
0.7362 -0.6889 0.0407 0 1.0000 -0.1742 0.5013 0.5127
0.4819 -0.7222 -0.8000 0 -6.4000 0.0962 0.3624 0.3168
0.3181 0.5082 1.6667 -0.5596 1.2000 0.0346 -0.0730 -0.0554
0.0367 0.6364 0.8878 0 0 -0.7922 -0.0323 -0.0659
0.5319 -15.0000 0.2676 0.7630 -Inf 0.5767 -0.1296 2.0361
0.6285 0.4459 0.0055 NaN 1.0000 0.0897 0.0051 0.7294
0.2664 1.5000 0.7692 Inf 0.0345 -0.6242 0.9130 1.0755
1.0929 0.5429 -0.0462 -0.1667 1.0000 3.8000 0.2618 0.9426
0.1665 0.2361 0.3803 9.5714 -0.8824 -0.0769 0.0388 -0.1921
0.4948 0.6216 1.7000 -0.7544 -1.7143 -0.3362 0.0374 0.4011
0.0695 -0.6667 0.1029 -0.0047 0.3400 0.1799 -0.1480 0.0041
0.2760 -0.1493 -0.0376 0.2468 0 -0.4798 0.5692 0.3966
0.8020 -0.3636 0 -0.1895 -0.0388 -0.2714 0.5421 0.5706
0.8096 0.7551 0.0551 -Inf -2.2500 0.0763 0.2739 -0.2762
0.1421 1.1600 0.5464 0.5082 1.0000 -0.2000 0.0794 1.7551
-0.1986 0.4615 -0.0847 -11.5000 4.9286 0.3684 2.1074 2.8250
0.0434 -3.8333 -0.1893 -22.0000 5.7500 0.2673 -0.7941 -0.3053
-1.1563 0.5541 0.1368 -0.6800 -0.0345 -0.2701 -0.6599 -0.8806
-1.1858 -1.0400 1.5072 0 0.1071 0.0216 -0.3361 -0.8307
0.5321 0.0833 1.1026 -0.2416 0 0.2484 0.0367 0.1931
0.3294 2.0000 -0.1111 -0.1825 7.6667 -0.3386 0.3154 0.4154
-0.0798 0.2679 0.3436 3.1944 -2.2222 -0.0109 -1.4390 -4.4595
-3.8451 1.3889 0.1172 1.0000 -1.7500 0.3240 0.5289 4.3000
Diffusion paramter specificity values (avg across subjs)
SI_a SI_ter SI_eta SI_sz SI_st SI_v1 SI_v2
SI_a =
0.5150
SI_ter =
0.2175
SI_eta =
0.7048
SI_sz =
-7.3500
SI_st =
0.0031
SI_v1 =
0.6641
SI_v2 =
0.7181
Bootstrap Specificity Indices and Confidence Intervals
N_samples = 1000 ; % If the bootstrapping has already been run, just load the file B.mat % Delete this file to rerun the bootstrap process... filename = fullfile(MAESpec02_pathstr,'diffusion','B.mat') ; %recalculatep = true ; recalculatep = false ; if (recalculatep || ~exist(filename,'file')) for k = N_samples:-1:1 % work backwards so that B(1000) is allocated first %- Sample subjects with replacement within each group if (k==1) % very fist sample always is the full data set sample = sbj_means ; % The new diffusion models sample2 = df_2 ; sample3 = df_3 ; sample4 = df_4 ; RT_all_sample = sRT_all ; RT_corr_sample = sRT_corr ; RTsd_all_sample = RTsd_all ; RTsd_corr_sample = RTsd_corr ; else idx = ceil(rand(1,N_sbj).*N_sbj) ; % Separate randomization for each model (is this necessary?) idx2 = ceil(rand(1,N_sbj).*N_sbj) ; idx3 = ceil(rand(1,N_sbj).*N_sbj) ; idx4 = ceil(rand(1,N_sbj).*N_sbj) ; sample = sbj_means(idx,:) ; sample2 = df_2(idx2,:) ; sample3 = df_3(idx3,:) ; sample4 = df_4(idx4,:) ; RT_all_sample = sRT_all(idx,:) ; RT_corr_sample = sRT_corr(idx,:) ; RTsd_all_sample = RTsd_all(idx,:) ; RTsd_corr_sample = RTsd_corr(idx,:) ; end % Pack this data away for later use % This is MODEL_1, which existed (without a subscript) prior to % October 2010. The indices were updated 2010-10-20 to accommodate % that now there are 11 periods instead of 8. Otherwise it's the % same. B(k).adpr = sample(:,dpr) ; B(k).edpr = sample(:,edpr_idx) ; B(k).hdpr = sample(:,hdpr_idx) ; B(k).a = sample(:,a) ; B(k).ter = sample(:,ter) ; B(k).eta = sample(:,eta) ; B(k).sz = sample(:,sz) ; B(k).st = sample(:,st) ; B(k).v1 = sample(:,v1) ; B(k).v2 = sample(:,v2) ; B(k).v_avg = (sample(:,v1) + sample(:,v2)) ./ 2 ; B(k).t1 = sample(:,ter) - (sample(:,st) ./ 2); B(k).t2 = sample(:,ter) + (sample(:,st) ./ 2); B(k).RT_all = RT_all_sample ; B(k).RT_corr = RT_corr_sample ; B(k).RTsd_all = RTsd_all_sample ; B(k).RTsd_corr = RTsd_corr_sample ; % Pack away the NEW diffusion model params (3 new models) % MODEL_2 -- added 2010-10-20 using MODEL_1 as a template B(k).a_2 = sample2(:,a_2) ; B(k).ter_2 = sample2(:,ter_2) ; B(k).eta_2 = sample2(:,eta_2) ; B(k).sz_2 = sample2(:,sz_2) ; B(k).st_2 = sample2(:,st_2) ; B(k).v1_2 = sample2(:,v1_2) ; B(k).v2_2 = sample2(:,v2_2) ; B(k).v_avg_2 = (sample2(:,v1_2) + sample2(:,v2_2)) ./ 2 ; B(k).t1_2 = sample2(:,ter_2) - (sample2(:,st_2) ./ 2); B(k).t2_2 = sample2(:,ter_2) + (sample2(:,st_2) ./ 2); % MODEL_3 -- added 2010-10-20 using MODEL_1 as a template B(k).a_3 = sample3(:,a_3) ; B(k).ter_3 = sample3(:,ter_3) ; B(k).eta_3 = sample3(:,eta_3) ; B(k).sz_3 = sample3(:,sz_3) ; B(k).st_3 = sample3(:,st_3) ; B(k).v1_3 = sample3(:,v1_3) ; B(k).v2_3 = sample3(:,v2_3) ; B(k).v_avg_3 = (sample3(:,v1_3) + sample3(:,v2_3)) ./ 2 ; B(k).t1_3 = sample3(:,ter_3) - repmat((sample3(:,st_3)./2),1,N_periods_total); B(k).t2_3 = sample3(:,ter_3) + repmat((sample3(:,st_3)./2),1,N_periods_total); % MODEL_4 -- added 2010-10-20 using MODEL_1 as a template B(k).a_4 = sample4(:,a_4) ; B(k).ter_4 = sample4(:,ter_4) ; B(k).eta_4 = sample4(:,eta_4) ; B(k).sz_4 = sample4(:,sz_4) ; B(k).st_4 = sample4(:,st_4) ; B(k).v1_4 = sample4(:,v1_4) ; B(k).v2_4 = sample4(:,v2_4) ; B(k).v_avg_4 = (sample4(:,v1_4) + sample4(:,v2_4)) ./ 2 ; B(k).t1_4 = sample4(:,ter_4) - (sample4(:,st_4) ./ 2); B(k).t2_4 = sample4(:,ter_4) + (sample4(:,st_4) ./ 2); % Pack away the specificity values for each smean = mean(sample,1) ; smean2 = mean(sample2,1) ; smean3 = mean(sample3,1) ; smean4 = mean(sample4,1) ; %tmp = smean(dpr) ; % obsolete code, subsumed within @specif_idx %B(k).adpr_SI = (tmp(5) - tmp(7))/ (tmp(5) - tmp(1)) ; B(k).adpr_SI = specif_idx(smean(dpr)) ; B(k).edpr_SI = specif_idx(smean(edpr_idx)) ; B(k).hdpr_SI = specif_idx(smean(hdpr_idx)) ; B(k).a_SI = specif_idx(smean(a)) ; B(k).ter_SI = specif_idx(smean(ter)) ; B(k).eta_SI = specif_idx(smean(eta)) ; B(k).sz_SI = specif_idx(smean(sz)) ; B(k).st_SI = specif_idx(smean(st)) ; B(k).v1_SI = specif_idx(smean(v1)) ; B(k).v2_SI = specif_idx(smean(v2)) ; B(k).v_avg_SI = specif_idx(mean(B(k).v_avg,1)) ; % the 'gain' is drift relative to dprime B(k).gain_SI = B(k).v_avg_SI - B(k).adpr_SI ; B(k).t1_SI = specif_idx(mean(B(k).t1,1)) ; B(k).t2_SI = specif_idx(mean(B(k).t2,1)) ; B(k).RT_all_SI = specif_idx(mean(B(k).RT_all,1)) ; B(k).RT_corr_SI = specif_idx(mean(B(k).RT_corr,1)) ; B(k).RTsd_all_SI = specif_idx(mean(B(k).RTsd_all,1)) ; B(k).RTsd_corr_SI = specif_idx(mean(B(k).RTsd_corr,1)) ; % The 3 new diffusion models. The fields that are fixed across all % periods for a particular model are commented out because no % specificity index can be calculated for them. 2010-10-21 % Model 2 -- this is the most constrainted model. Only drift rates % vary across periods, so there is only SI for drift %B(k).a_SI_2 = specif_idx(smean2(a_2)) ; %B(k).ter_SI_2 = specif_idx(smean2(ter_2)) ; %B(k).eta_SI_2 = specif_idx(smean2(eta_2)) ; %B(k).sz_SI_2 = specif_idx(smean2(sz_2)) ; %B(k).st_SI_2 = specif_idx(smean2(st_2)) ; B(k).v1_SI_2 = specif_idx(smean2(v1_2)) ; B(k).v2_SI_2 = specif_idx(smean2(v2_2)) ; B(k).v_avg_SI_2 = specif_idx(mean(B(k).v_avg_2,1)) ; B(k).gain_SI_2 = B(k).v_avg_SI_2 - B(k).adpr_SI ; %B(k).t1_SI_2 = specif_idx(mean(B(k).t1_2,1)) ; %B(k).t2_SI_2 = specif_idx(mean(B(k).t2_2,1)) ; % Model 3 -- Ter and drift vary and hence have SIs %B(k).a_SI_3 = specif_idx(smean3(a_3)) ; B(k).ter_SI_3 = specif_idx(smean3(ter_3)) ; %B(k).eta_SI_3 = specif_idx(smean3(eta_3)) ; %B(k).sz_SI_3 = specif_idx(smean3(sz_3)) ; %B(k).st_SI_3 = specif_idx(smean3(st_3)) ; B(k).v1_SI_3 = specif_idx(smean3(v1_3)) ; B(k).v2_SI_3 = specif_idx(smean3(v2_3)) ; B(k).v_avg_SI_3 = specif_idx(mean(B(k).v_avg_3,1)) ; B(k).gain_SI_3 = B(k).v_avg_SI_3 - B(k).adpr_SI ; %B(k).t1_SI_3 = specif_idx(mean(B(k).t1_3,1)) ; %B(k).t2_SI_3 = specif_idx(mean(B(k).t2_3,1)) ; % Model 4 -- Ter, st, and drift vary. Eta, a, and sz are fixed. %B(k).a_SI_4 = specif_idx(smean4(a_4)) ; B(k).ter_SI_4 = specif_idx(smean4(ter_4)) ; %B(k).eta_SI_4 = specif_idx(smean4(eta_4)) ; %B(k).sz_SI_4 = specif_idx(smean4(sz_4)) ; B(k).st_SI_4 = specif_idx(smean4(st_4)) ; B(k).v1_SI_4 = specif_idx(smean4(v1_4)) ; B(k).v2_SI_4 = specif_idx(smean4(v2_4)) ; B(k).v_avg_SI_4 = specif_idx(mean(B(k).v_avg_4,1)) ; B(k).gain_SI_4 = B(k).v_avg_SI_4 - B(k).adpr_SI ; B(k).t1_SI_4 = specif_idx(mean(B(k).t1_4,1)) ; B(k).t2_SI_4 = specif_idx(mean(B(k).t2_4,1)) ; % Learning indices ********************************************* B(k).adpr_LI = learn_idx(smean(dpr)) ; B(k).edpr_LI = learn_idx(smean(edpr_idx)) ; B(k).hdpr_LI = learn_idx(smean(hdpr_idx)) ; B(k).a_LI = learn_idx(smean(a)) ; B(k).ter_LI = learn_idx(smean(ter)) ; B(k).eta_LI = learn_idx(smean(eta)) ; B(k).sz_LI = learn_idx(smean(sz)) ; B(k).st_LI = learn_idx(smean(st)) ; B(k).v1_LI = learn_idx(smean(v1)) ; B(k).v2_LI = learn_idx(smean(v2)) ; B(k).v_avg_LI = learn_idx(mean(B(k).v_avg,1)) ; B(k).gain_LI = B(k).v_avg_LI - B(k).adpr_LI ; B(k).t1_LI = learn_idx(mean(B(k).t1,1)) ; B(k).t2_LI = learn_idx(mean(B(k).t2,1)) ; B(k).RT_all_LI = learn_idx(mean(B(k).RT_all,1)) ; B(k).RT_corr_LI = learn_idx(mean(B(k).RT_corr,1)) ; B(k).RTsd_all_LI = learn_idx(mean(B(k).RTsd_all,1)) ; B(k).RTsd_corr_LI = learn_idx(mean(B(k).RTsd_corr,1)) ; % The 3 new diffusion models. The fields that are fixed across all % periods for a particular model are commented out because no % learning index can be calculated for them. 2010-10-21 % Model 2 -- this is the most constrainted model. Only drift rates % vary across periods, so there is only LI for drift %B(k).a_LI_2 = learn_idx(smean2(a_2)) ; %B(k).ter_LI_2 = learn_idx(smean2(ter_2)) ; %B(k).eta_LI_2 = learn_idx(smean2(eta_2)) ; %B(k).sz_LI_2 = learn_idx(smean2(sz_2)) ; %B(k).st_LI_2 = learn_idx(smean2(st_2)) ; B(k).v1_LI_2 = learn_idx(smean2(v1_2)) ; B(k).v2_LI_2 = learn_idx(smean2(v2_2)) ; B(k).v_avg_LI_2 = learn_idx(mean(B(k).v_avg_2,1)) ; B(k).gain_LI_2 = B(k).v_avg_LI_2 - B(k).adpr_LI ; %B(k).t1_LI_2 = learn_idx(mean(B(k).t1_2,1)) ; %B(k).t2_LI_2 = learn_idx(mean(B(k).t2_2,1)) ; % Model 3 -- Ter and drift vary and hence have LIs %B(k).a_LI_3 = learn_idx(smean3(a_3)) ; B(k).ter_LI_3 = learn_idx(smean3(ter_3)) ; %B(k).eta_LI_3 = learn_idx(smean3(eta_3)) ; %B(k).sz_LI_3 = learn_idx(smean3(sz_3)) ; %B(k).st_LI_3 = learn_idx(smean3(st_3)) ; B(k).v1_LI_3 = learn_idx(smean3(v1_3)) ; B(k).v2_LI_3 = learn_idx(smean3(v2_3)) ; B(k).v_avg_LI_3 = learn_idx(mean(B(k).v_avg_3,1)) ; B(k).gain_LI_3 = B(k).v_avg_LI_3 - B(k).adpr_LI ; %B(k).t1_LI_3 = learn_idx(mean(B(k).t1_3,1)) ; %B(k).t2_LI_3 = learn_idx(mean(B(k).t2_3,1)) ; % Model 4 -- Ter, st, and drift vary. Eta, a, and sz are fixed. %B(k).a_LI_4 = learn_idx(smean4(a_4)) ; B(k).ter_LI_4 = learn_idx(smean4(ter_4)) ; %B(k).eta_LI_4 = learn_idx(smean4(eta_4)) ; %B(k).sz_LI_4 = learn_idx(smean4(sz_4)) ; B(k).st_LI_4 = learn_idx(smean4(st_4)) ; B(k).v1_LI_4 = learn_idx(smean4(v1_4)) ; B(k).v2_LI_4 = learn_idx(smean4(v2_4)) ; B(k).v_avg_LI_4 = learn_idx(mean(B(k).v_avg_4,1)) ; B(k).gain_LI_4 = B(k).v_avg_LI_4 - B(k).adpr_LI ; B(k).t1_LI_4 = learn_idx(mean(B(k).t1_4,1)) ; B(k).t2_LI_4 = learn_idx(mean(B(k).t2_4,1)) ; end clear tmp; % Save the structure B for later save(filename,'B') ; else fprintf('load %s \n\n',filename) ; load(filename) ; end
load /Users/apetrov/a/r/w/work/MLExper/MAESpec02/diffusion/B.mat
B(1) contains the SIs and LIs on the full data set
B(1)
ans =
adpr: [27x11 double]
edpr: [27x11 double]
hdpr: [27x11 double]
a: [27x11 double]
ter: [27x11 double]
eta: [27x11 double]
sz: [27x11 double]
st: [27x11 double]
v1: [27x11 double]
v2: [27x11 double]
v_avg: [27x11 double]
t1: [27x11 double]
t2: [27x11 double]
RT_all: [27x11 double]
RT_corr: [27x11 double]
RTsd_all: [27x11 double]
RTsd_corr: [27x11 double]
a_2: [27x1 double]
ter_2: [27x1 double]
eta_2: [27x1 double]
sz_2: [27x1 double]
st_2: [27x1 double]
v1_2: [27x11 double]
v2_2: [27x11 double]
v_avg_2: [27x11 double]
t1_2: [27x1 double]
t2_2: [27x1 double]
a_3: [27x1 double]
ter_3: [27x11 double]
eta_3: [27x1 double]
sz_3: [27x1 double]
st_3: [27x1 double]
v1_3: [27x11 double]
v2_3: [27x11 double]
v_avg_3: [27x11 double]
t1_3: [27x11 double]
t2_3: [27x11 double]
a_4: [27x1 double]
ter_4: [27x11 double]
eta_4: [27x1 double]
sz_4: [27x1 double]
st_4: [27x11 double]
v1_4: [27x11 double]
v2_4: [27x11 double]
v_avg_4: [27x11 double]
t1_4: [27x11 double]
t2_4: [27x11 double]
adpr_SI: 0.6034
edpr_SI: 0.5534
hdpr_SI: 0.6588
a_SI: 0.5150
ter_SI: 0.2175
eta_SI: 0.7048
sz_SI: -7.3500
st_SI: 0.0031
v1_SI: 0.6641
v2_SI: 0.7181
v_avg_SI: 0.6851
gain_SI: 0.0816
t1_SI: 0.8909
t2_SI: 0.1250
RT_all_SI: 0.3660
RT_corr_SI: 0.3621
RTsd_all_SI: 0.9392
RTsd_corr_SI: 0.9614
v1_SI_2: 0.6230
v2_SI_2: 0.6793
v_avg_SI_2: 0.6427
gain_SI_2: 0.0393
ter_SI_3: 0.3880
v1_SI_3: 0.5503
v2_SI_3: 0.6685
v_avg_SI_3: 0.5950
gain_SI_3: -0.0085
ter_SI_4: 0.3174
st_SI_4: 0.2316
v1_SI_4: 0.5642
v2_SI_4: 0.6636
v_avg_SI_4: 0.6029
gain_SI_4: -5.3980e-04
t1_SI_4: 0.4890
t2_SI_4: 0.2831
adpr_LI: 0.5478
edpr_LI: 0.5554
hdpr_LI: 0.6525
a_LI: -0.0909
ter_LI: -0.2040
eta_LI: 0.3710
sz_LI: -0.0326
st_LI: -0.5582
v1_LI: 0.9573
v2_LI: 1.0469
v_avg_LI: 0.9902
gain_LI: 0.4424
t1_LI: -0.0682
t2_LI: -0.2809
RT_all_LI: -0.2734
RT_corr_LI: -0.2734
RTsd_all_LI: -0.1072
RTsd_corr_LI: -0.1236
v1_LI_2: 1.0282
v2_LI_2: 0.9050
v_avg_LI_2: 0.9814
gain_LI_2: 0.4336
ter_LI_3: -0.2040
v1_LI_3: 0.7731
v2_LI_3: 0.7936
v_avg_LI_3: 0.7807
gain_LI_3: 0.2329
ter_LI_4: -0.2588
st_LI_4: -0.6169
v1_LI_4: 0.6760
v2_LI_4: 0.7411
v_avg_LI_4: 0.6999
gain_LI_4: 0.1521
t1_LI_4: -0.1198
t2_LI_4: -0.3370
Average over all N_samples to find more stable SI values
MODEL_1, which prior to Oct 2010 was the only model -- the saturated model
SI_adpr_b = [B.adpr_SI]' ; SI_edpr_b = [B.edpr_SI]' ; SI_hdpr_b = [B.hdpr_SI]' ; SI_a_b = [B.a_SI]' ; SI_ter_b = [B.ter_SI]' ; SI_eta_b = [B.eta_SI]' ; SI_sz_b = [B.sz_SI]' ; SI_st_b = [B.st_SI]' ; SI_v1_b = [B.v1_SI]' ; SI_v2_b = [B.v2_SI]' ; % The three new models (2 thourgh 4), as of 2010-10-20: SI_v1_b_2 = [B.v1_SI_2]' ; SI_v2_b_2 = [B.v2_SI_2]' ; SI_v1_b_3 = [B.v1_SI_3]' ; SI_v2_b_3 = [B.v2_SI_3]' ; SI_ter_b_3 = [B.ter_SI_3]' ; SI_v1_b_4 = [B.v1_SI_4]' ; SI_v2_b_4 = [B.v2_SI_4]' ; SI_ter_b_4 = [B.ter_SI_4]' ; SI_st_b_4 = [B.st_SI_4]' ; % Special averaged drift rate SI_v_avg_b = [B.v_avg_SI]' ; SI_v_avg_b_2 = [B.v_avg_SI_2]' ; SI_v_avg_b_3 = [B.v_avg_SI_3]' ; SI_v_avg_b_4 = [B.v_avg_SI_4]' ; % Special "gain" over traditional dprime analysis SI_gain_b = [B.gain_SI]' ; SI_gain_b_2 = [B.gain_SI_2]' ; SI_gain_b_3 = [B.gain_SI_3]' ; SI_gain_b_4 = [B.gain_SI_4]' ; % Define Ter/St interaction variables SI_t1_b = [B.t1_SI]' ; SI_t2_b = [B.t2_SI]' ; SI_t1_b_4 = [B.t1_SI_4]' ; SI_t2_b_4 = [B.t2_SI_4]' ; SI_RT_all_b = [B.RT_all_SI]' ; SI_RT_corr_b = [B.RT_corr_SI]' ; SI_RTsd_all_b = [B.RTsd_all_SI]' ; SI_RTsd_corr_b = [B.RTsd_corr_SI]' ;
Average over all N_samples to find more stable LI values
MODEL_1, which prior to Oct 2010 was the only model -- the saturated model
LI_adpr_b = [B.adpr_LI]' ; LI_edpr_b = [B.edpr_LI]' ; LI_hdpr_b = [B.hdpr_LI]' ; LI_a_b = [B.a_LI]' ; LI_ter_b = [B.ter_LI]' ; LI_eta_b = [B.eta_LI]' ; LI_sz_b = [B.sz_LI]' ; LI_st_b = [B.st_LI]' ; LI_v1_b = [B.v1_LI]' ; LI_v2_b = [B.v2_LI]' ; % The three new models (2 thourgh 4), as of 2010-10-20: LI_v1_b_2 = [B.v1_LI_2]' ; LI_v2_b_2 = [B.v2_LI_2]' ; LI_v1_b_3 = [B.v1_LI_3]' ; LI_v2_b_3 = [B.v2_LI_3]' ; LI_ter_b_3 = [B.ter_LI_3]' ; LI_v1_b_4 = [B.v1_LI_4]' ; LI_v2_b_4 = [B.v2_LI_4]' ; LI_ter_b_4 = [B.ter_LI_4]' ; LI_st_b_4 = [B.st_LI_4]' ; % Special averaged drift rate LI_v_avg_b = [B.v_avg_LI]' ; LI_v_avg_b_2 = [B.v_avg_LI_2]' ; LI_v_avg_b_3 = [B.v_avg_LI_3]' ; LI_v_avg_b_4 = [B.v_avg_LI_4]' ; % Special "gain" over traditional dprime analysis LI_gain_b = [B.gain_LI]' ; LI_gain_b_2 = [B.gain_LI_2]' ; LI_gain_b_3 = [B.gain_LI_3]' ; LI_gain_b_4 = [B.gain_LI_4]' ; % Define Ter/St interaction variables LI_t1_b = [B.t1_LI]' ; LI_t2_b = [B.t2_LI]' ; LI_t1_b_4 = [B.t1_LI_4]' ; LI_t2_b_4 = [B.t2_LI_4]' ; LI_RT_all_b = [B.RT_all_LI]' ; LI_RT_corr_b = [B.RT_corr_LI]' ; LI_RTsd_all_b = [B.RTsd_all_LI]' ; LI_RTsd_corr_b = [B.RTsd_corr_LI]' ;
Collect distribution statistics of the 11-point profiles
D-prime
adpr_gr = NaN(N_samples, N_periods_total) ; edpr_gr = NaN(N_samples, N_periods_total) ; hdpr_gr = NaN(N_samples, N_periods_total) ; % MODEL_1 a_gr = NaN(N_samples, N_periods_total) ; ter_gr = NaN(N_samples, N_periods_total) ; eta_gr = NaN(N_samples, N_periods_total) ; sz_gr = NaN(N_samples, N_periods_total) ; st_gr = NaN(N_samples, N_periods_total) ; v1_gr = NaN(N_samples, N_periods_total) ; v2_gr = NaN(N_samples, N_periods_total) ; % Special averaged drift rate v_avg_gr = NaN(N_samples,N_periods_total) ; t1_gr = NaN(N_samples,N_periods_total) ; t2_gr = NaN(N_samples,N_periods_total) ; RT_all_gr = NaN(N_samples,N_periods_total) ; RT_corr_gr = NaN(N_samples,N_periods_total) ; RTsd_all_gr = NaN(N_samples,N_periods_total) ; RTsd_corr_gr = NaN(N_samples,N_periods_total) ; % The three new models (featuring 11-point profiles) % Model 2: a_gr_2 = NaN(N_samples, 1) ; ter_gr_2 = NaN(N_samples, 1) ; eta_gr_2 = NaN(N_samples, 1) ; sz_gr_2 = NaN(N_samples, 1) ; st_gr_2 = NaN(N_samples, 1) ; v1_gr_2 = NaN(N_samples, N_periods_total) ; v2_gr_2 = NaN(N_samples, N_periods_total) ; % Special averaged drift rate v_avg_gr_2 = NaN(N_samples,N_periods_total) ; % Model 3: a_gr_3 = NaN(N_samples, 1) ; eta_gr_3 = NaN(N_samples, 1) ; sz_gr_3 = NaN(N_samples, 1) ; st_gr_3 = NaN(N_samples, 1) ; ter_gr_3 = NaN(N_samples, N_periods_total) ; v1_gr_3 = NaN(N_samples, N_periods_total) ; v2_gr_3 = NaN(N_samples, N_periods_total) ; % Special averaged drift rate v_avg_gr_3 = NaN(N_samples,N_periods_total) ; % Model 4: a_gr_4 = NaN(N_samples, 1) ; eta_gr_4 = NaN(N_samples, 1) ; sz_gr_4 = NaN(N_samples, 1) ; ter_gr_4 = NaN(N_samples, N_periods_total) ; st_gr_4 = NaN(N_samples, N_periods_total) ; v1_gr_4 = NaN(N_samples, N_periods_total) ; v2_gr_4 = NaN(N_samples, N_periods_total) ; % Special averaged drift rate v_avg_gr_4 = NaN(N_samples,N_periods_total) ; % Calculate averages across the bootstrap samples for k=1:N_samples % Eleven-point curves for each group % D-prime adpr_gr(k,:) = mean(B(k).adpr,1) ; edpr_gr(k,:) = mean(B(k).edpr,1) ; hdpr_gr(k,:) = mean(B(k).hdpr,1) ; % Model 1 a_gr(k,:) = mean(B(k).a,1) ; ter_gr(k,:) = mean(B(k).ter,1) ; eta_gr(k,:) = mean(B(k).eta,1) ; sz_gr(k,:) = mean(B(k).sz,1) ; st_gr(k,:) = mean(B(k).st,1) ; v1_gr(k,:) = mean(B(k).v1,1) ; v2_gr(k,:) = mean(B(k).v2,1) ; v_avg_gr(k,:) = mean(B(k).v_avg,1) ; t1_gr(k,:) = mean(B(k).t1,1) ; t2_gr(k,:) = mean(B(k).t2,1) ; RT_all_gr(k,:) = mean(B(k).RT_all,1) ; RT_corr_gr(k,:) = mean(B(k).RT_corr,1) ; RTsd_all_gr(k,:) = mean(B(k).RTsd_all,1) ; RTsd_corr_gr(k,:) = mean(B(k).RTsd_corr,1) ; % The three new models % Model 2 a_gr_2(k,:) = mean(B(k).a_2,1) ; ter_gr_2(k,:) = mean(B(k).ter_2,1) ; eta_gr_2(k,:) = mean(B(k).eta_2,1) ; sz_gr_2(k,:) = mean(B(k).sz_2,1) ; st_gr_2(k,:) = mean(B(k).st_2,1) ; v1_gr_2(k,:) = mean(B(k).v1_2,1) ; v2_gr_2(k,:) = mean(B(k).v2_2,1) ; v_avg_gr_2(k,:) = mean(B(k).v_avg_2,1) ; % Model 3 a_gr_3(k,:) = mean(B(k).a_3,1) ; eta_gr_3(k,:) = mean(B(k).eta_3,1) ; sz_gr_3(k,:) = mean(B(k).sz_3,1) ; st_gr_3(k,:) = mean(B(k).st_3,1) ; ter_gr_3(k,:) = mean(B(k).ter_3,1) ; v1_gr_3(k,:) = mean(B(k).v1_3,1) ; v2_gr_3(k,:) = mean(B(k).v2_3,1) ; v_avg_gr_3(k,:) = mean(B(k).v_avg_3,1) ; % Model 4 a_gr_4(k,:) = mean(B(k).a_4,1) ; eta_gr_4(k,:) = mean(B(k).eta_4,1) ; sz_gr_4(k,:) = mean(B(k).sz_4,1) ; ter_gr_4(k,:) = mean(B(k).ter_4,1) ; st_gr_4(k,:) = mean(B(k).st_4,1) ; v1_gr_4(k,:) = mean(B(k).v1_4,1) ; v2_gr_4(k,:) = mean(B(k).v2_4,1) ; v_avg_gr_4(k,:) = mean(B(k).v_avg_4,1) ; end
Plot bootstrap dprime curve and SI value for single dprime
This is old code (prior to 2010-10-21) -- the aggregate-dprime version of panel a in Figure~2.
% Find BS confidence interval bs_adpr = prctile(adpr_gr,[10 50 90],1) ; figure % Single dprime curve M = 3.5 ; plot(1:N_periods_total, bs_adpr(2,:),'b.-') ; axis([1 N_periods_total 0 M]) ; grid on ; set(gca,'xtick',([1:N_periods_total]),... 'xticklabel',[],'ytick',(0:ceil(M))) ; title(sprintf('Bootstrap dprime for %d samples', N_samples)) ; ylabel('dprime'); xlabel(sprintf('SI = %0.3f', (median(SI_adpr_b)))); errorbar1(1:N_periods_total,bs_adpr(2,:),bs_adpr(2,:) - bs_adpr(1,:),... bs_adpr(3,:) - bs_adpr(2,:)) ;
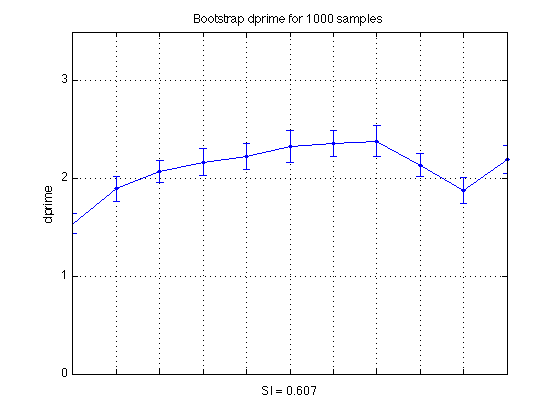
Bootstrap-based dprime learning curves
Plot bootstrap dprime curves and SI values for easy and hard dprime Panel A in Figure 2 in the PBR manuscript submitted 2010-08-03. Updated to 11 time periods 2010-10-21 NOTE: This was abandoned in the revision of 2010-11-17. The revised figure does not use any bootstrap values. It is based on the empirical means and the within-subject CIs instead -- see the above section titled "Export data for the six-plot empirical figure, 2010-11-17".
% Find BS confidence interval bs_edpr = prctile(edpr_gr,[10 50 90],1) ; bs_hdpr = prctile(hdpr_gr,[10 50 90],1) ; figure % Easy and difficult dprimes plot(1:N_periods_total, bs_edpr(2,:),'b.-') ; errorbar1(1:N_periods_total,bs_edpr(2,:),bs_edpr(2,:) - bs_edpr(1,:),... bs_edpr(3,:) - bs_edpr(2,:),'b') ; hold on plot(1:N_periods_total, bs_hdpr(2,:),'r.-') ; errorbar1(1:N_periods_total,bs_hdpr(2,:),bs_hdpr(2,:) - bs_hdpr(1,:),... bs_hdpr(3,:) - bs_hdpr(2,:), 'r') ; axis([1 N_periods_total 0 M]) ; grid on ; set(gca,'xtick',([1:N_periods_total]),... 'xticklabel',[],'ytick',(0:ceil(M))) ; title(sprintf('Bootstrap dprime for %d samples', N_samples)) ; ylabel('dprime'); xlabel(sprintf('SI(e) = %0.3f ; SI(d) = %0.3f', median(SI_edpr_b), median(SI_hdpr_b)));
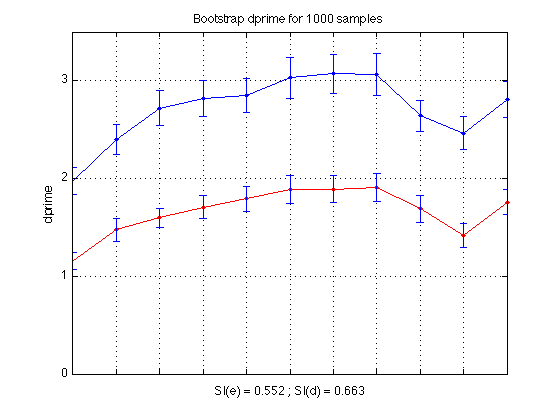
Plot the resulting diffusion param bootstrap curves with SI written below
Old (prior to Oct 2010) ==> All for the saturated MODEL 1. Updated for 11 periods, 2010-10-21.
% Find BS confidence interval bs_a = prctile(a_gr,[10 50 90],1) ; bs_ter = prctile(ter_gr,[10 50 90],1) ; bs_eta = prctile(eta_gr,[10 50 90],1) ; bs_sz = prctile(sz_gr,[10 50 90],1) ; bs_st = prctile(st_gr,[10 50 90],1) ; bs_v1 = prctile(v1_gr,[10 50 90],1) ; bs_v2 = prctile(v2_gr,[10 50 90],1) ; bs_v_avg = prctile(v_avg_gr,[10 50 90],1) ; figure subplot(3,2,1) ; % a plot(1:N_periods_total, bs_a(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_a(2,:),bs_a(2,:) - bs_a(1,:),... bs_a(3,:) - bs_a(2,:)) ; title('Boundary a'); xlabel(sprintf('SI = %0.3f', median(SI_a_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0.10 0.15]) ; grid on ; subplot(3,2,3) ; % ter plot(1:N_periods_total, bs_ter(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_ter(2,:)*1000,(bs_ter(2,:) - bs_ter(1,:))*1000,... (bs_ter(3,:) - bs_ter(2,:))*1000) ; title('Nondecision Ter'); xlabel(sprintf('SI = %0.3f', median(SI_ter_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 300 600]) ; grid on ; subplot(3,2,5) ; % eta plot(1:N_periods_total, bs_eta(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_eta(2,:),bs_eta(2,:) - bs_eta(1,:),... bs_eta(3,:) - bs_eta(2,:)) ; title('SD Drift in eta'); xlabel(sprintf('SI = %0.3f', median(SI_eta_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 0.35]) ; grid on ; subplot(3,2,2) ; % sz plot(1:N_periods_total, bs_sz(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_sz(2,:),bs_sz(2,:) - bs_sz(1,:),... bs_sz(3,:) - bs_sz(2,:)) ; title('Range in z, sz'); xlabel(sprintf('SI = %0.3f', median(SI_sz_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 0.07]) ; grid on ; subplot(3,2,4) ; % st plot(1:N_periods_total, bs_st(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_st(2,:)*1000,(bs_st(2,:) - bs_st(1,:))*1000,... (bs_st(3,:) - bs_st(2,:))*1000) ; title('Range in nondecision, st'); xlabel(sprintf('SI = %0.3f', median(SI_st_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 300]) ; grid on ; % v1 subplot(3,2,6) ; plot(1:N_periods_total, bs_v1(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_v1(2,:),bs_v1(2,:) - bs_v1(1,:),... bs_v1(3,:) - bs_v1(2,:)) ; title('Drift rates'); xlabel(sprintf('SI(v1) = %0.3f ; v = %0.3f ; SI(v2) = %0.3f',... median(SI_v1_b),median(SI_v_avg_b),median(SI_v2_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); hold on % v2 plot(1:N_periods_total, bs_v2(2,:), 'r.-'); errorbar1(1:N_periods_total,bs_v2(2,:),bs_v2(2,:) - bs_v2(1,:),... bs_v2(3,:) - bs_v2(2,:)) ; axis([1 N_periods_total 0 0.5]) ; grid on ; % v avg hold on plot(1:N_periods_total, bs_v_avg(2,:), 'g.-'); errorbar1(1:N_periods_total,bs_v_avg(2,:),bs_v_avg(2,:) - bs_v_avg(1,:),... bs_v_avg(3,:) - bs_v_avg(2,:)) ; hold off
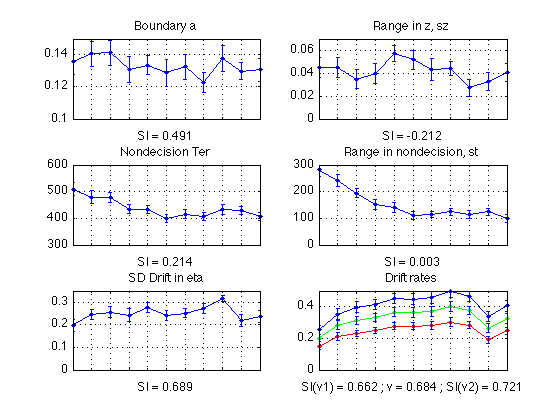
Ter analysis
Old (prior to Oct 2010) ==> All for the saturated MODEL 1. Updated for 11 periods, 2010-10-21.
bs_t1 = prctile(t1_gr,[10 50 90],1) ; bs_t2 = prctile(t2_gr,[10 50 90],1) ; figure subplot(2,2,1) ; % ter plot(1:N_periods_total, bs_ter(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_ter(2,:)*1000,(bs_ter(2,:) - bs_ter(1,:))*1000,... (bs_ter(3,:) - bs_ter(2,:))*1000) ; title('Nondecision Ter'); xlabel(sprintf('SI = %0.3f', median(SI_ter_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 300 600]) ; grid on ; subplot(2,2,2) ; % st plot(1:N_periods_total, bs_st(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_st(2,:)*1000,(bs_st(2,:) - bs_st(1,:))*1000,... (bs_st(3,:) - bs_st(2,:))*1000) ; title('Range in nondecision, st'); xlabel(sprintf('SI = %0.3f', median(SI_st_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 300]) ; grid on ; subplot(2,2,3) ; % Ter - st/2 = t1 plot(1:N_periods_total, bs_t1(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_t1(2,:)*1000,(bs_t1(2,:) - bs_t1(1,:))*1000,... (bs_t1(3,:) - bs_t1(2,:))*1000) ; title('Minimum non-decision time t1'); xlabel(sprintf('SI = %0.3f', median(SI_t1_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 300 600]) ; grid on ; subplot(2,2,4) ; % Ter + st/2 = t2 plot(1:N_periods_total, bs_t2(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_t2(2,:)*1000,(bs_t2(2,:) - bs_t2(1,:))*1000,... (bs_t2(3,:) - bs_t2(2,:))*1000) ; title('Maximum non--decision time t2'); xlabel(sprintf('SI = %0.3f', median(SI_t2_b))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 400 700]) ; grid on ;
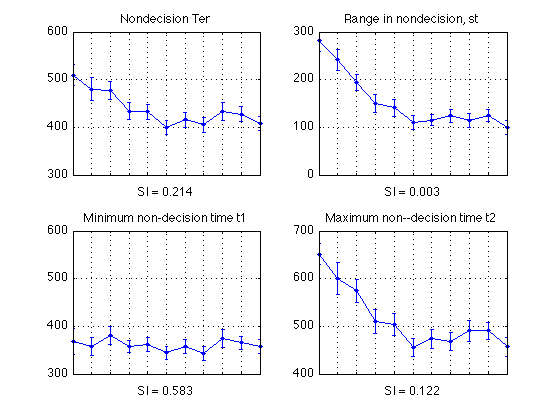
New diffusion models (2-4)
This code was added by Nick 2010-10-20
% Find BS confidence interval % Model 2 bs_a_2 = prctile(a_gr_2,[10 50 90],1) ; bs_ter_2 = prctile(ter_gr_2,[10 50 90],1) ; bs_eta_2 = prctile(eta_gr_2,[10 50 90],1) ; bs_sz_2 = prctile(sz_gr_2,[10 50 90],1) ; bs_st_2 = prctile(st_gr_2,[10 50 90],1) ; bs_v1_2 = prctile(v1_gr_2,[10 50 90],1) ; bs_v2_2 = prctile(v2_gr_2,[10 50 90],1) ; bs_v_avg_2 = prctile(v_avg_gr_2,[10 50 90],1) ; % Model 3 bs_a_3 = prctile(a_gr_3,[10 50 90],1) ; bs_ter_3 = prctile(ter_gr_3,[10 50 90],1) ; bs_eta_3 = prctile(eta_gr_3,[10 50 90],1) ; bs_sz_3 = prctile(sz_gr_3,[10 50 90],1) ; bs_st_3 = prctile(st_gr_3,[10 50 90],1) ; bs_v1_3 = prctile(v1_gr_3,[10 50 90],1) ; bs_v2_3 = prctile(v2_gr_3,[10 50 90],1) ; bs_v_avg_3 = prctile(v_avg_gr_3,[10 50 90],1) ; % Model 4 bs_a_4 = prctile(a_gr_4,[10 50 90],1) ; bs_ter_4 = prctile(ter_gr_4,[10 50 90],1) ; bs_eta_4 = prctile(eta_gr_4,[10 50 90],1) ; bs_sz_4 = prctile(sz_gr_4,[10 50 90],1) ; bs_st_4 = prctile(st_gr_4,[10 50 90],1) ; bs_v1_4 = prctile(v1_gr_4,[10 50 90],1) ; bs_v2_4 = prctile(v2_gr_4,[10 50 90],1) ; bs_v_avg_4 = prctile(v_avg_gr_4,[10 50 90],1) ; figure % Plot model 2 (only drift rates apply here) % v1 subplot(3,3,1) ; plot(1:N_periods_total, bs_v1_2(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_v1_2(2,:),bs_v1_2(2,:) - bs_v1_2(1,:),... bs_v1_2(3,:) - bs_v1_2(2,:)) ; title('Drift rates (Model 2)'); xlabel(sprintf('SI(v1) = %0.3f ; v = %0.3f ; SI(v2) = %0.3f',... median(SI_v1_b_2),median(SI_v_avg_b_2),median(SI_v2_b_2))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); hold on % v2 plot(1:N_periods_total, bs_v2_2(2,:), 'r.-'); errorbar1(1:N_periods_total,bs_v2_2(2,:),bs_v2_2(2,:) - bs_v2_2(1,:),... bs_v2_2(3,:) - bs_v2_2(2,:)) ; axis([1 N_periods_total 0 0.5]) ; grid on ; % v avg hold on plot(1:N_periods_total, bs_v_avg_2(2,:), 'g.-'); errorbar1(1:N_periods_total,bs_v_avg_2(2,:),bs_v_avg_2(2,:) - bs_v_avg_2(1,:),... bs_v_avg_2(3,:) - bs_v_avg_2(2,:)) ; hold off % Plot model 3 % v1 subplot(3,3,4) ; plot(1:N_periods_total, bs_v1_3(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_v1_3(2,:),bs_v1_3(2,:) - bs_v1_3(1,:),... bs_v1_3(3,:) - bs_v1_3(2,:)) ; title('Drift rates (Model 3)'); xlabel(sprintf('SI(v1) = %0.3f ; v = %0.3f ; SI(v2) = %0.3f',... median(SI_v1_b_3),median(SI_v_avg_b_3),median(SI_v2_b_3))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); hold on % v2 plot(1:N_periods_total, bs_v2_3(2,:), 'r.-'); errorbar1(1:N_periods_total,bs_v2_3(2,:),bs_v2_3(2,:) - bs_v2_3(1,:),... bs_v2_3(3,:) - bs_v2_3(2,:)) ; axis([1 N_periods_total 0 0.5]) ; grid on ; % v avg hold on plot(1:N_periods_total, bs_v_avg_3(2,:), 'g.-'); errorbar1(1:N_periods_total,bs_v_avg_3(2,:),bs_v_avg_3(2,:) - bs_v_avg_3(1,:),... bs_v_avg_3(3,:) - bs_v_avg_3(2,:)) ; hold off % ter subplot(3,3,5) ; plot(1:N_periods_total, bs_ter_3(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_ter_3(2,:)*1000,(bs_ter_3(2,:) - bs_ter_3(1,:))*1000,... (bs_ter_3(3,:) - bs_ter_3(2,:))*1000) ; title('Nondec Ter (Model 3)'); xlabel(sprintf('SI = %0.3f', median(SI_ter_b_3))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 300 600]) ; grid on ; % Plot model 4 subplot(3,3,7) ; % v1 plot(1:N_periods_total, bs_v1_4(2,:), 'b.-'); errorbar1(1:N_periods_total,bs_v1_4(2,:),bs_v1_4(2,:) - bs_v1_4(1,:),... bs_v1_4(3,:) - bs_v1_4(2,:)) ; title('Drift rates (Model 4)'); xlabel(sprintf('SI(v1) = %0.3f ; v = %0.3f ; SI(v2) = %0.3f',... median(SI_v1_b_4),median(SI_v_avg_b_4),median(SI_v2_b_4))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); hold on % v2 plot(1:N_periods_total, bs_v2_4(2,:), 'r.-'); errorbar1(1:N_periods_total,bs_v2_4(2,:),bs_v2_4(2,:) - bs_v2_4(1,:),... bs_v2_4(3,:) - bs_v2_4(2,:)) ; axis([1 N_periods_total 0 0.5]) ; grid on ; % v avg hold on plot(1:N_periods_total, bs_v_avg_4(2,:), 'g.-'); errorbar1(1:N_periods_total,bs_v_avg_4(2,:),bs_v_avg_4(2,:) - bs_v_avg_4(1,:),... bs_v_avg_4(3,:) - bs_v_avg_4(2,:)) ; hold off % ter subplot(3,3,8) ; plot(1:N_periods_total, bs_ter_4(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_ter_4(2,:)*1000,(bs_ter_4(2,:) - bs_ter_4(1,:))*1000,... (bs_ter_4(3,:) - bs_ter_4(2,:))*1000) ; title('Nondec Ter (Model 4)'); xlabel(sprintf('SI = %0.3f', median(SI_ter_b_4))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 300 600]) ; grid on ; % st subplot(3,3,9) ; plot(1:N_periods_total, bs_st_4(2,:)*1000, 'b.-'); errorbar1(1:N_periods_total,bs_st_4(2,:)*1000,(bs_st_4(2,:) - bs_st_4(1,:))*1000,... (bs_st_4(3,:) - bs_st_4(2,:))*1000) ; title('Rng in nondec, st (Model 4)'); xlabel(sprintf('SI = %0.3f', median(SI_st_b_4))); set(gca,'xtick',([1:N_periods_total]),'xticklabel',[]); axis([1 N_periods_total 0 300]) ; grid on ;
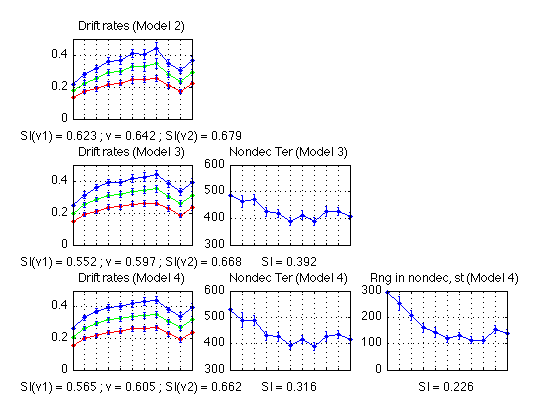
Plot Bootstrap RT and RT std (sbjs averaged together)
Old (prior to Oct 2010). Based on bootstrapping the raw RTs Updated for 11 periods, 2010-10-21.
% Find BS confidence interval bs_RT_all = prctile(RT_all_gr,[10 50 90],1) ; bs_RT_corr = prctile(RT_corr_gr,[10 50 90],1) ; bs_RTsd_all = prctile(RTsd_all_gr,[10 50 90],1) ; bs_RTsd_corr = prctile(RTsd_corr_gr,[10 50 90],1) ; % All RTs subplot(2,2,1) ; plot(1:N_periods_total, bs_RT_all(2,:), 'b.-') ; axis([1 N_periods_total 400 800]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT For All Trials') ; ylabel('msec') ; xlabel(sprintf('SI = %0.3f', median(SI_RT_all_b))); errorbar1(1:N_periods_total,bs_RT_all(2,:),bs_RT_all(2,:) - bs_RT_all(1,:),... bs_RT_all(3,:) - bs_RT_all(2,:)) ; % Correct RTs subplot(2,2,3) ; plot(1:N_periods_total, bs_RT_corr(2,:), 'b.-') ; axis([1 N_periods_total 400 800]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT For Correct Trials') ; ylabel('msec') ; xlabel(sprintf('SI = %0.3f', median(SI_RT_corr_b))); errorbar1(1:N_periods_total,bs_RT_corr(2,:),bs_RT_corr(2,:) - bs_RT_corr(1,:),... bs_RT_corr(3,:) - bs_RT_corr(2,:)) ; % All RT std subplot(2,2,2) ; plot(1:N_periods_total, bs_RTsd_all(2,:), 'b.-') ; axis([1 N_periods_total 200 400]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT std For All Trials') ; ylabel('msec') ; xlabel(sprintf('SI = %0.3f', median(SI_RTsd_all_b))); errorbar1(1:N_periods_total,bs_RTsd_all(2,:),bs_RTsd_all(2,:) - bs_RTsd_all(1,:),... bs_RTsd_all(3,:) - bs_RTsd_all(2,:)) ; % Correct RT std subplot(2,2,4) ; plot(1:N_periods_total, bs_RTsd_corr(2,:), 'b.-') ; axis([1 N_periods_total 200 400]) ; grid on ; set(gca,'xtick',([1:N_periods_total])); title('RT std For All Trials') ; ylabel('msec') ; xlabel(sprintf('SI = %0.3f', median(SI_RTsd_corr_b))); errorbar1(1:N_periods_total,bs_RTsd_corr(2,:),bs_RTsd_corr(2,:) - bs_RTsd_corr(1,:),... bs_RTsd_corr(3,:) - bs_RTsd_corr(2,:)) ;
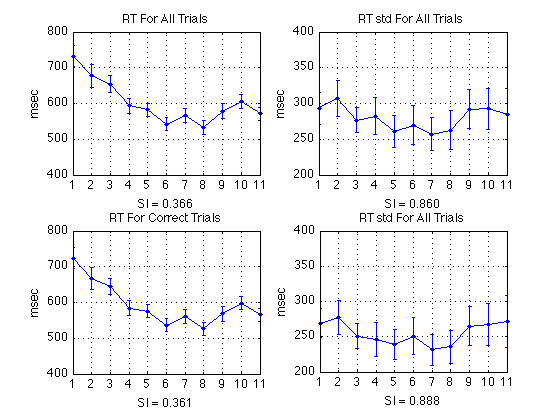
Raw bootstrap percentiles
fprintf('\n---------------------------------------------\n') fprintf('Raw data statistics\n'); fprintf('---------------------------------------------\n') fprintf('Row 1 = Lower bound (10th percentile)\n'); fprintf('Row 2 = Median (50th percentile)\n'); fprintf('Row 3 = Upper bound (90th percentile)\n'); fprintf('\nColumns = Eight "blocks"\n'); fprintf('---------------------------------------------\n') % All dprime bs_adpr % Easy dprime bs_edpr % Hard dprime bs_hdpr % All RTs bs_RT_all % Correct RTs bs_RT_corr % All RTs std bs_RTsd_all % Correct RTs std bs_RTsd_corr % Diffusion params bs_a bs_ter bs_eta bs_sz bs_st bs_v1 bs_v2 bs_v_avg bs_t1 bs_t2 % Diffusion Model 2 bs_a_2 bs_ter_2 bs_eta_2 bs_sz_2 bs_st_2 bs_v1_2 bs_v2_2 bs_v_avg_2 % Diffusion Model 3 bs_a_3 bs_ter_3 bs_eta_3 bs_sz_3 bs_st_3 bs_v1_3 bs_v2_3 bs_v_avg_3 % Diffusion Model 4 bs_a_4 bs_ter_4 bs_eta_4 bs_sz_4 bs_st_4 bs_v1_4 bs_v2_4 bs_v_avg_4
---------------------------------------------
Raw data statistics
---------------------------------------------
Row 1 = Lower bound (10th percentile)
Row 2 = Median (50th percentile)
Row 3 = Upper bound (90th percentile)
Columns = Eight "blocks"
---------------------------------------------
bs_adpr =
1.4367 1.7657 1.9649 2.0288 2.0930 2.1692 2.2237 2.2244 2.0168 1.7400 2.0548
1.5393 1.8934 2.0750 2.1690 2.2242 2.3303 2.3597 2.3820 2.1384 1.8744 2.1955
1.6471 2.0170 2.1983 2.2967 2.3647 2.4955 2.4971 2.5308 2.2709 2.0035 2.3351
bs_edpr =
1.8416 2.2488 2.5407 2.6314 2.6777 2.8155 2.8753 2.8518 2.4783 2.2962 2.6301
1.9759 2.4028 2.7216 2.8181 2.8518 3.0297 3.0738 3.0680 2.6409 2.4637 2.8137
2.1140 2.5631 2.9010 2.9966 3.0467 3.2535 3.2572 3.2808 2.7956 2.6332 2.9891
bs_hdpr =
1.0725 1.3564 1.4994 1.5867 1.6649 1.7461 1.7577 1.7639 1.5554 1.2910 1.6334
1.1569 1.4745 1.5988 1.7069 1.7911 1.8907 1.8925 1.9093 1.6917 1.4161 1.7602
1.2422 1.5894 1.7003 1.8349 1.9276 2.0382 2.0234 2.0503 1.8419 1.5266 1.8968
bs_RT_all =
702.8218 645.6204 631.5556 571.7940 564.8542 524.1273 546.3194 513.1389 558.1574 587.0741 553.0926
732.6065 677.7894 654.3472 593.2269 582.7639 541.9398 566.0463 532.2870 578.9444 606.0231 570.7685
762.6759 711.8171 676.2477 616.0093 601.2917 562.3727 587.5278 553.8032 599.7593 626.0949 591.5154
bs_RT_corr =
695.2593 636.7037 620.9815 563.0602 558.7292 519.5162 540.0347 507.7153 547.2500 579.5162 548.0556
723.9306 667.6319 643.8310 583.6319 575.9213 536.9028 559.6991 526.2569 567.9259 598.2755 565.1049
753.1528 701.1250 664.9120 605.8449 593.9699 557.1806 580.1713 547.1412 588.9722 618.3218 585.8179
bs_RTsd_all =
271.2380 281.7221 258.8286 256.0314 238.9274 242.1797 234.1187 235.2190 265.0464 263.9384 249.0007
293.4101 306.9057 276.3318 282.0221 261.0090 269.4625 257.1695 262.6774 292.2507 292.3318 285.2090
320.2189 340.8848 298.3680 308.6940 282.6419 297.4763 282.2577 288.1292 319.2524 321.1733 323.1568
bs_RTsd_corr =
248.6649 253.7839 233.0209 222.2371 217.0721 224.7152 209.2572 212.2859 237.1992 237.8397 234.8352
269.2033 277.5831 250.7157 246.2273 238.3459 250.7240 231.5255 235.5596 264.8405 267.1865 271.7885
292.2763 306.2322 272.4131 269.4260 259.1715 277.2152 255.8084 258.1075 290.1545 298.2473 311.3206
bs_a =
0.1278 0.1328 0.1336 0.1229 0.1276 0.1206 0.1249 0.1166 0.1295 0.1247 0.1234
0.1358 0.1407 0.1414 0.1311 0.1337 0.1288 0.1327 0.1229 0.1376 0.1300 0.1306
0.1436 0.1495 0.1489 0.1398 0.1415 0.1367 0.1405 0.1306 0.1450 0.1355 0.1379
bs_ter =
0.4869 0.4555 0.4579 0.4167 0.4169 0.3851 0.3995 0.3900 0.4134 0.4127 0.3925
0.5098 0.4797 0.4774 0.4340 0.4328 0.4001 0.4155 0.4051 0.4325 0.4279 0.4078
0.5340 0.5061 0.4970 0.4520 0.4517 0.4184 0.4348 0.4245 0.4538 0.4465 0.4279
bs_eta =
0.1714 0.2216 0.2334 0.2146 0.2560 0.2213 0.2305 0.2485 0.3013 0.1939 0.2101
0.1982 0.2456 0.2567 0.2426 0.2767 0.2435 0.2504 0.2712 0.3159 0.2205 0.2372
0.2245 0.2699 0.2773 0.2684 0.2962 0.2656 0.2685 0.2934 0.3291 0.2449 0.2624
bs_sz =
0.0360 0.0365 0.0263 0.0311 0.0500 0.0437 0.0339 0.0378 0.0200 0.0252 0.0330
0.0448 0.0453 0.0350 0.0400 0.0574 0.0518 0.0436 0.0439 0.0274 0.0329 0.0407
0.0539 0.0549 0.0442 0.0498 0.0637 0.0596 0.0530 0.0500 0.0367 0.0415 0.0482
bs_st =
0.2587 0.2194 0.1766 0.1314 0.1223 0.0964 0.1036 0.1109 0.1003 0.1119 0.0842
0.2820 0.2415 0.1937 0.1505 0.1406 0.1104 0.1150 0.1242 0.1151 0.1250 0.0997
0.3049 0.2637 0.2117 0.1694 0.1598 0.1245 0.1269 0.1383 0.1295 0.1376 0.1142
bs_v1 =
0.2272 0.3192 0.3670 0.3841 0.4231 0.4146 0.4185 0.4592 0.4355 0.3047 0.3689
0.2553 0.3539 0.3987 0.4144 0.4543 0.4489 0.4601 0.4989 0.4678 0.3373 0.4059
0.2845 0.3877 0.4307 0.4463 0.4882 0.4824 0.4975 0.5329 0.4988 0.3704 0.4396
bs_v2 =
0.1314 0.1886 0.2140 0.2289 0.2553 0.2543 0.2554 0.2771 0.2630 0.1707 0.2234
0.1480 0.2133 0.2312 0.2501 0.2782 0.2784 0.2808 0.3035 0.2839 0.1926 0.2469
0.1666 0.2358 0.2500 0.2720 0.3014 0.3017 0.3030 0.3273 0.3091 0.2125 0.2691
bs_v_avg =
0.1797 0.2547 0.2908 0.3068 0.3391 0.3341 0.3375 0.3684 0.3507 0.2382 0.2968
0.2015 0.2838 0.3150 0.3321 0.3660 0.3636 0.3705 0.4016 0.3764 0.2647 0.3265
0.2253 0.3119 0.3401 0.3581 0.3942 0.3910 0.4004 0.4301 0.4021 0.2909 0.3545
bs_t1 =
0.3409 0.3392 0.3620 0.3453 0.3478 0.3308 0.3435 0.3287 0.3555 0.3515 0.3439
0.3686 0.3584 0.3805 0.3583 0.3620 0.3446 0.3583 0.3431 0.3748 0.3654 0.3587
0.3986 0.3812 0.3990 0.3722 0.3791 0.3616 0.3757 0.3602 0.3972 0.3825 0.3761
bs_t2 =
0.6298 0.5672 0.5503 0.4842 0.4810 0.4362 0.4545 0.4483 0.4683 0.4723 0.4377
0.6514 0.6006 0.5744 0.5095 0.5038 0.4557 0.4735 0.4676 0.4905 0.4905 0.4576
0.6731 0.6344 0.5982 0.5341 0.5277 0.4778 0.4946 0.4902 0.5142 0.5114 0.4810
bs_a_2 =
0.1336
0.1385
0.1433
bs_ter_2 =
0.3928
0.4076
0.4269
bs_eta_2 =
0.1972
0.2163
0.2347
bs_sz_2 =
0.0310
0.0379
0.0445
bs_st_2 =
0.1512
0.1710
0.1920
bs_v1_2 =
0.2013 0.2571 0.2904 0.3395 0.3430 0.3816 0.3735 0.4021 0.3269 0.2825 0.3416
0.2174 0.2752 0.3127 0.3615 0.3662 0.4082 0.4053 0.4395 0.3490 0.3016 0.3659
0.2342 0.2940 0.3363 0.3844 0.3910 0.4358 0.4381 0.4829 0.3724 0.3230 0.3882
bs_v2_2 =
0.1214 0.1571 0.1743 0.2014 0.2071 0.2244 0.2251 0.2332 0.1919 0.1573 0.2044
0.1331 0.1711 0.1878 0.2164 0.2244 0.2442 0.2444 0.2540 0.2087 0.1717 0.2205
0.1449 0.1848 0.2016 0.2309 0.2423 0.2629 0.2633 0.2769 0.2284 0.1871 0.2373
bs_v_avg_2 =
0.1615 0.2077 0.2324 0.2706 0.2752 0.3044 0.2993 0.3181 0.2614 0.2198 0.2744
0.1753 0.2231 0.2503 0.2891 0.2954 0.3263 0.3252 0.3468 0.2782 0.2367 0.2931
0.1891 0.2391 0.2680 0.3074 0.3159 0.3489 0.3504 0.3794 0.2987 0.2546 0.3120
bs_a_3 =
0.1319
0.1368
0.1416
bs_ter_3 =
0.4628 0.4406 0.4506 0.4084 0.4042 0.3744 0.3965 0.3730 0.4059 0.4124 0.3940
0.4865 0.4627 0.4696 0.4243 0.4193 0.3890 0.4123 0.3877 0.4243 0.4264 0.4075
0.5124 0.4872 0.4889 0.4426 0.4363 0.4076 0.4302 0.4057 0.4426 0.4439 0.4249
bs_eta_3 =
0.2194
0.2374
0.2556
bs_sz_3 =
0.0416
0.0496
0.0570
bs_st_3 =
0.1279
0.1406
0.1525
bs_v1_3 =
0.2317 0.2936 0.3402 0.3692 0.3733 0.3917 0.3981 0.4127 0.3608 0.3112 0.3659
0.2473 0.3124 0.3590 0.3893 0.3917 0.4160 0.4241 0.4386 0.3817 0.3322 0.3898
0.2626 0.3287 0.3782 0.4095 0.4126 0.4407 0.4523 0.4638 0.4023 0.3556 0.4141
bs_v2_3 =
0.1379 0.1760 0.1990 0.2196 0.2243 0.2384 0.2423 0.2447 0.2116 0.1701 0.2183
0.1466 0.1900 0.2099 0.2350 0.2390 0.2553 0.2591 0.2624 0.2292 0.1848 0.2332
0.1549 0.2037 0.2210 0.2506 0.2553 0.2736 0.2760 0.2799 0.2481 0.2000 0.2475
bs_v_avg_3 =
0.1852 0.2360 0.2701 0.2947 0.2994 0.3167 0.3204 0.3297 0.2892 0.2407 0.2926
0.1969 0.2511 0.2843 0.3120 0.3153 0.3353 0.3416 0.3502 0.3049 0.2587 0.3120
0.2082 0.2659 0.2986 0.3296 0.3340 0.3560 0.3640 0.3715 0.3241 0.2771 0.3296
bs_a_4 =
0.1277
0.1327
0.1377
bs_ter_4 =
0.5038 0.4627 0.4681 0.4136 0.4095 0.3776 0.4002 0.3759 0.4081 0.4185 0.3994
0.5286 0.4897 0.4867 0.4307 0.4257 0.3924 0.4164 0.3908 0.4272 0.4341 0.4145
0.5529 0.5168 0.5054 0.4482 0.4432 0.4098 0.4336 0.4078 0.4456 0.4509 0.4322
bs_eta_4 =
0.2291
0.2471
0.2638
bs_sz_4 =
0.0355
0.0432
0.0508
bs_st_4 =
0.2733 0.2241 0.1874 0.1429 0.1250 0.1061 0.1172 0.0990 0.1009 0.1392 0.1202
0.2962 0.2510 0.2060 0.1596 0.1429 0.1187 0.1292 0.1124 0.1136 0.1549 0.1377
0.3186 0.2784 0.2266 0.1759 0.1598 0.1311 0.1415 0.1289 0.1261 0.1705 0.1564
bs_v1_4 =
0.2480 0.3131 0.3565 0.3784 0.3811 0.3964 0.4056 0.4191 0.3625 0.3200 0.3754
0.2636 0.3291 0.3731 0.3968 0.4017 0.4185 0.4320 0.4413 0.3803 0.3411 0.3984
0.2788 0.3453 0.3906 0.4165 0.4252 0.4412 0.4609 0.4663 0.3977 0.3626 0.4216
bs_v2_4 =
0.1434 0.1843 0.2065 0.2229 0.2297 0.2437 0.2452 0.2506 0.2130 0.1758 0.2226
0.1533 0.1967 0.2172 0.2370 0.2455 0.2604 0.2609 0.2665 0.2291 0.1916 0.2364
0.1624 0.2099 0.2291 0.2530 0.2622 0.2784 0.2787 0.2846 0.2455 0.2073 0.2513
bs_v_avg_4 =
0.1961 0.2496 0.2826 0.3009 0.3054 0.3212 0.3261 0.3352 0.2901 0.2481 0.2994
0.2082 0.2629 0.2953 0.3173 0.3233 0.3395 0.3465 0.3539 0.3050 0.2662 0.3174
0.2198 0.2773 0.3091 0.3343 0.3427 0.3591 0.3694 0.3753 0.3193 0.2837 0.3353
Descriptive statistics for the on-line supplement, 2010-07-14
Calculate an omnibus CI80 for each variable, pooling the CIs over the 11 time periods. Assume the weights are inversely proportional to the square root of the number of blocks in the corresponding period. Work directly with the difference b/n 90th and 10th percentile -- it is proportional to the standard deviation (assuming normal distribution).
Updated for 11 time periods 2010-10-20.
blocks_per_period = [4 4 4 4 4 4 4 4 1 4 3] ; assert(length(blocks_per_period)==N_periods_total) ; std_per_period = 1./sqrt(blocks_per_period) ; % passed to combine_measurements combined_CI = @(bs) (combine_measurements((bs(3,:)-bs(1,:))/2,std_per_period)) ; CI80_summary.adpr = combined_CI(bs_adpr) ; CI80_summary.edpr = combined_CI(bs_edpr) ; CI80_summary.hdpr = combined_CI(bs_hdpr) ; CI80_summary.RT_all = combined_CI(bs_RT_all) ; CI80_summary.RT_corr = combined_CI(bs_RT_corr) ; CI80_summary.RTsd_all = combined_CI(bs_RTsd_all) ; CI80_summary.RTsd_corr = combined_CI(bs_RTsd_corr) ; CI80_summary.a = combined_CI(bs_a) ; CI80_summary.ter = combined_CI(bs_ter) ; CI80_summary.eta = combined_CI(bs_eta) ; CI80_summary.sz = combined_CI(bs_sz) ; CI80_summary.st = combined_CI(bs_st) ; CI80_summary.v1 = combined_CI(bs_v1) ; CI80_summary.v2 = combined_CI(bs_v2) ; CI80_summary.v_avg = combined_CI(bs_v_avg) ; CI80_summary.t1 = combined_CI(bs_t1) ; CI80_summary.t2 = combined_CI(bs_t2) ; % Model 2, added 2010-10-20 CI80_summary.a_2 = combined_CI(bs_a_2) ; CI80_summary.ter_2 = combined_CI(bs_ter_2) ; CI80_summary.eta_2 = combined_CI(bs_eta_2) ; CI80_summary.sz_2 = combined_CI(bs_sz_2) ; CI80_summary.st_2 = combined_CI(bs_st_2) ; CI80_summary.v1_2 = combined_CI(bs_v1_2) ; CI80_summary.v2_2 = combined_CI(bs_v2_2) ; CI80_summary.v_avg_2 = combined_CI(bs_v_avg_2) ; % Model 3, added 2010-10-20 CI80_summary.a_3 = combined_CI(bs_a_3) ; CI80_summary.ter_3 = combined_CI(bs_ter_3) ; CI80_summary.eta_3 = combined_CI(bs_eta_3) ; CI80_summary.sz_3 = combined_CI(bs_sz_3) ; CI80_summary.st_3 = combined_CI(bs_st_3) ; CI80_summary.v1_3 = combined_CI(bs_v1_3) ; CI80_summary.v2_3 = combined_CI(bs_v2_3) ; CI80_summary.v_avg_3 = combined_CI(bs_v_avg_3) ; % Model 4, added 2010-10-20 CI80_summary.a_4 = combined_CI(bs_a_4) ; CI80_summary.ter_4 = combined_CI(bs_ter_4) ; CI80_summary.eta_4 = combined_CI(bs_eta_4) ; CI80_summary.sz_4 = combined_CI(bs_sz_4) ; CI80_summary.st_4 = combined_CI(bs_st_4) ; CI80_summary.v1_4 = combined_CI(bs_v1_4) ; CI80_summary.v2_4 = combined_CI(bs_v2_4) ; CI80_summary.v_avg_4 = combined_CI(bs_v_avg_4) %%%%% The grand_means will be used in the ANOVAs below %%%%% 'Grand' in the sense that it takes all 27 original subjects [B(1)] %%%%% and averages across all of them. So, these are the group averaged %%%%% profiles, still varying across 11 time periods. grand_means.adpr = mean(B(1).adpr) ; % B(1) is the full group grand_means.edpr = mean(B(1).edpr) ; grand_means.hdpr = mean(B(1).hdpr) ; grand_means.RT_all = mean(B(1).RT_all) ; grand_means.RT_corr = mean(B(1).RT_corr) ; grand_means.RTsd_all = mean(B(1).RTsd_all) ; grand_means.RTsd_corr = mean(B(1).RTsd_corr) ; grand_means.a = mean(B(1).a) ; grand_means.ter = mean(B(1).ter) ; grand_means.eta = mean(B(1).eta) ; grand_means.sz = mean(B(1).sz) ; grand_means.st = mean(B(1).st) ; grand_means.v1 = mean(B(1).v1) ; grand_means.v2 = mean(B(1).v2) ; grand_means.v_avg = mean(B(1).v_avg) ; grand_means.t1 = mean(B(1).t1) ; grand_means.t2 = mean(B(1).t2) ; % Model 2 grand_means.a_2 = mean(B(1).a_2) ; grand_means.ter_2 = mean(B(1).ter_2) ; grand_means.eta_2 = mean(B(1).eta_2) ; grand_means.sz_2 = mean(B(1).sz_2) ; grand_means.st_2 = mean(B(1).st_2) ; grand_means.v1_2 = mean(B(1).v1_2) ; grand_means.v2_2 = mean(B(1).v2_2) ; grand_means.v_avg_2 = mean(B(1).v_avg_2) ; % Model 3 grand_means.a_3 = mean(B(1).a_3) ; grand_means.ter_3 = mean(B(1).ter_3) ; grand_means.eta_3 = mean(B(1).eta_3) ; grand_means.sz_3 = mean(B(1).sz_3) ; grand_means.st_3 = mean(B(1).st_3) ; grand_means.v1_3 = mean(B(1).v1_3) ; grand_means.v2_3 = mean(B(1).v2_3) ; grand_means.v_avg_3 = mean(B(1).v_avg_3) ; % Model 4 grand_means.a_4 = mean(B(1).a_4) ; grand_means.ter_4 = mean(B(1).ter_4) ; grand_means.eta_4 = mean(B(1).eta_4) ; grand_means.sz_4 = mean(B(1).sz_4) ; grand_means.st_4 = mean(B(1).st_4) ; grand_means.v1_4 = mean(B(1).v1_4) ; grand_means.v2_4 = mean(B(1).v2_4) ; grand_means.v_avg_4 = mean(B(1).v_avg_4) ; grand_means.t1_4 = mean(B(1).t1_4) ; grand_means.t2_4 = mean(B(1).t2_4)
CI80_summary =
adpr: 0.1339
edpr: 0.1808
hdpr: 0.1232
RT_all: 22.4876
RT_corr: 21.9528
RTsd_all: 26.3405
RTsd_corr: 25.0312
a: 0.0075
ter: 0.0190
eta: 0.0232
sz: 0.0083
st: 0.0168
v1: 0.0336
v2: 0.0220
v_avg: 0.0274
t1: 0.0178
t2: 0.0231
a_2: 0.0049
ter_2: 0.0170
eta_2: 0.0187
sz_2: 0.0068
st_2: 0.0204
v1_2: 0.0247
v2_2: 0.0164
v_avg_2: 0.0201
a_3: 0.0049
ter_3: 0.0182
eta_3: 0.0181
sz_3: 0.0077
st_3: 0.0123
v1_3: 0.0215
v2_3: 0.0147
v_avg_3: 0.0174
a_4: 0.0050
ter_4: 0.0186
eta_4: 0.0173
sz_4: 0.0076
st_4: 0.0175
v1_4: 0.0206
v2_4: 0.0147
v_avg_4: 0.0170
grand_means =
adpr: [1.5379 1.8925 2.0733 2.1644 2.2251 2.3292 2.3588 2.3805 2.1424 1.8721 2.1925]
edpr: [1.9742 2.4024 2.7152 2.8128 2.8620 3.0312 3.0682 3.0706 2.6388 2.4638 2.8112]
hdpr: [1.1553 1.4729 1.5972 1.7069 1.7923 1.8890 1.8887 1.9091 1.6944 1.4125 1.7599]
RT_all: [734.2176 679.6898 655.1343 594.0093 583.6713 543.3009 566.7315 533.5093 579.3333 606.9676 571.8580]
RT_corr: [725.7454 670.0093 644.6019 584.3611 576.8380 538.4167 560.4676 527.3519 568.2222 599.1944 566.4568]
RTsd_all: [295.8909 311.1253 278.7040 284.2011 262.0796 271.3128 258.4584 264.1647 293.6426 293.9606 288.1520]
RTsd_corr: [270.4425 280.6370 252.7658 247.9733 239.4832 252.3316 232.7824 237.0102 265.4384 269.1505 274.5641]
a: [0.1361 0.1415 0.1418 0.1316 0.1346 0.1292 0.1331 0.1238 0.1380 0.1301 0.1311]
ter: [0.5100 0.4799 0.4776 0.4338 0.4334 0.4006 0.4164 0.4060 0.4333 0.4286 0.4090]
eta: [0.1974 0.2446 0.2556 0.2412 0.2765 0.2433 0.2500 0.2707 0.3159 0.2191 0.2370]
sz: [0.0454 0.0458 0.0353 0.0407 0.0570 0.0521 0.0440 0.0440 0.0284 0.0331 0.0411]
st: [0.2827 0.2416 0.1946 0.1511 0.1416 0.1107 0.1158 0.1249 0.1156 0.1254 0.0996]
v1: [0.2541 0.3517 0.3980 0.4133 0.4546 0.4477 0.4588 0.4973 0.4682 0.3358 0.4050]
v2: [0.1477 0.2118 0.2311 0.2497 0.2776 0.2769 0.2798 0.3023 0.2855 0.1913 0.2461]
v_avg: [0.2009 0.2818 0.3145 0.3315 0.3661 0.3623 0.3693 0.3998 0.3768 0.2635 0.3256]
t1: [0.3686 0.3591 0.3803 0.3582 0.3626 0.3453 0.3585 0.3435 0.3755 0.3659 0.3592]
t2: [0.6514 0.6007 0.5749 0.5094 0.5042 0.4559 0.4743 0.4684 0.4911 0.4913 0.4588]
a_2: 0.1386
ter_2: 0.4083
eta_2: 0.2151
sz_2: 0.0380
st_2: 0.1720
v1_2: [0.2166 0.2741 0.3110 0.3601 0.3654 0.4069 0.4032 0.4393 0.3478 0.3006 0.3639]
v2_2: [0.1326 0.1701 0.1871 0.2150 0.2231 0.2427 0.2430 0.2526 0.2086 0.1711 0.2197]
v_avg_2: [0.1746 0.2221 0.2490 0.2875 0.2942 0.3248 0.3231 0.3459 0.2782 0.2358 0.2918]
a_3: 0.1370
ter_3: [0.4880 0.4633 0.4698 0.4250 0.4200 0.3901 0.4128 0.3885 0.4240 0.4271 0.4081]
eta_3: 0.2381
sz_3: 0.0499
st_3: 0.1404
v1_3: [0.2475 0.3122 0.3591 0.3894 0.3931 0.4165 0.4261 0.4388 0.3817 0.3335 0.3904]
v2_3: [0.1464 0.1899 0.2100 0.2355 0.2400 0.2559 0.2593 0.2626 0.2297 0.1849 0.2336]
v_avg_3: [0.1969 0.2511 0.2846 0.3124 0.3165 0.3362 0.3427 0.3507 0.3057 0.2592 0.3120]
a_4: 0.1326
ter_4: [0.5284 0.4901 0.4868 0.4311 0.4262 0.3935 0.4170 0.3917 0.4274 0.4351 0.4154]
eta_4: 0.2460
sz_4: 0.0429
st_4: [0.2955 0.2509 0.2065 0.1597 0.1424 0.1190 0.1291 0.1132 0.1136 0.1554 0.1380]
v1_4: [0.2636 0.3295 0.3737 0.3974 0.4021 0.4184 0.4321 0.4419 0.3803 0.3413 0.3986]
v2_4: [0.1532 0.1972 0.2177 0.2376 0.2458 0.2606 0.2614 0.2667 0.2290 0.1914 0.2367]
v_avg_4: [0.2084 0.2634 0.2957 0.3175 0.3239 0.3395 0.3468 0.3543 0.3047 0.2663 0.3176]
t1_4: [0.3806 0.3647 0.3835 0.3513 0.3550 0.3340 0.3525 0.3351 0.3706 0.3574 0.3464]
t2_4: [0.6762 0.6155 0.5900 0.5110 0.4974 0.4530 0.4816 0.4483 0.4842 0.5128 0.4844]
Specificity-index summary table
Anonymous helper functions added by Alex, 2010-06-29 Updated for 11 time periods by Nick, 2010-10-20
SI_stats_prctile = @(b) (prctile(b,[10 50 90])) ; % [Q10 median Q90] SI_stats_prctile_w_CI = @(p) ([p, (p(3)-p(1))/2]) ; % [Q10 median Q90, CI80] SI_stats_helper = @(b) ... (sprintf('%8.4f',max(-99,[SI_stats_prctile_w_CI(SI_stats_prctile(b)),mean(b),b(1)]))) ; fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('All dprim: %s\n', SI_stats_helper(SI_adpr_b)) ; fprintf('Easy dprim: %s\n', SI_stats_helper(SI_edpr_b)) ; fprintf('Hard dprim: %s\n', SI_stats_helper(SI_hdpr_b)) ; fprintf('RT_all: %s\n', SI_stats_helper(SI_RT_all_b)) ; fprintf('RT_correct: %s\n', SI_stats_helper(SI_RT_corr_b)) ; fprintf('RT_all std: %s\n', SI_stats_helper(SI_RTsd_all_b)) ; fprintf('RT_corr std: %s\n', SI_stats_helper(SI_RTsd_corr_b)) ; fprintf('---------------------------------------------------------------\n') fprintf('Param a: %s\n', SI_stats_helper(SI_a_b)) ; fprintf('Param ter: %s\n', SI_stats_helper(SI_ter_b)) ; fprintf('Param eta: %s\n', SI_stats_helper(SI_eta_b)) ; fprintf('Param sz: %s\n', SI_stats_helper(SI_sz_b)) ; fprintf('Param st: %s\n', SI_stats_helper(SI_st_b)) ; fprintf('Param v1 easy: %s\n', SI_stats_helper(SI_v1_b)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(SI_v2_b)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(SI_v_avg_b)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(SI_gain_b)) ; fprintf('t1=Ter-st/2: %s\n', SI_stats_helper(SI_t1_b)) ; fprintf('t2=Ter+st/2: %s\n', SI_stats_helper(SI_t2_b)) ; fprintf('---------------------------------------------------------------\n') fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') % Model 2 fprintf('\n\n==== MODEL 2 (Does not do well in terms of BIC) =====\n') ; fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('Param v1 easy: %s\n', SI_stats_helper(SI_v1_b_2)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(SI_v2_b_2)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(SI_v_avg_b_2)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(SI_gain_b_2)) ; fprintf('---------------------------------------------------------------\n') fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') % Model 3 fprintf('\n\n==== MODEL 3 (Does not do well in terms of BIC) =====\n') ; fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('Param ter: %s\n', SI_stats_helper(SI_ter_b_3)) ; fprintf('Param v1 easy: %s\n', SI_stats_helper(SI_v1_b_3)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(SI_v2_b_3)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(SI_v_avg_b_3)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(SI_gain_b_3)) ; fprintf('---------------------------------------------------------------\n') fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') % Model 4 fprintf('\n\n==== MODEL 4 =====\n') ; fprintf('Specif. indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('Param ter: %s\n', SI_stats_helper(SI_ter_b_4)) ; fprintf('Param st: %s\n', SI_stats_helper(SI_st_b_4)) ; fprintf('Param v1 easy: %s\n', SI_stats_helper(SI_v1_b_4)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(SI_v2_b_4)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(SI_v_avg_b_4)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(SI_gain_b_4)) ; fprintf('t1=Ter-st/2: %s\n', SI_stats_helper(SI_t1_b_4)) ; fprintf('t2=Ter+st/2: %s\n', SI_stats_helper(SI_t2_b_4)) ; fprintf('---------------------------------------------------------------\n') fprintf('Specif. indices Q10 median Q90 CI80 mean full\n')
Specif. indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- All dprim: 0.5012 0.6071 0.7042 0.1015 0.6027 0.6034 Easy dprim: 0.4567 0.5516 0.6339 0.0886 0.5494 0.5534 Hard dprim: 0.5419 0.6633 0.7755 0.1168 0.6603 0.6588 RT_all: 0.3005 0.3657 0.4416 0.0706 0.3691 0.3660 RT_correct: 0.2984 0.3614 0.4334 0.0675 0.3648 0.3621 RT_all std: 0.2699 0.8595 2.3351 1.0326 1.4897 0.9392 RT_corr std: 0.3044 0.8877 2.3615 1.0285 2.8970 0.9614 --------------------------------------------------------------- Param a: -0.0429 0.4912 1.2673 0.6551-99.0000 0.5150 Param ter: 0.1346 0.2138 0.3256 0.0955 0.2227 0.2175 Param eta: 0.3564 0.6892 1.2426 0.4431 0.7206 0.7048 Param sz: -5.1097 -0.2123 5.1570 5.1333 Inf -7.3500 Param st: -0.0771 0.0027 0.0793 0.0782 0.0015 0.0031 Param v1 easy: 0.5744 0.6619 0.7664 0.0960 0.6656 0.6641 Param v2 hard: 0.6324 0.7207 0.8158 0.0917 0.7228 0.7181 --------------------------------------------------------------- Avg v: 0.6013 0.6843 0.7817 0.0902 0.6877 0.6851 Gain=v-adpr: -0.0261 0.0789 0.2064 0.1162 0.0850 0.0816 t1=Ter-st/2: -1.6945 0.5832 2.4610 2.0778 0.5658 0.8909 t2=Ter+st/2: 0.0582 0.1224 0.1926 0.0672 0.1241 0.1250 --------------------------------------------------------------- Specif. indices Q10 median Q90 CI80 mean full ==== MODEL 2 (Does not do well in terms of BIC) ===== Specif. indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- Param v1 easy: 0.5469 0.6232 0.6888 0.0710 0.6204 0.6230 Param v2 hard: 0.5878 0.6788 0.7694 0.0908 0.6788 0.6793 --------------------------------------------------------------- Avg v: 0.5687 0.6417 0.7121 0.0717 0.6407 0.6427 Gain=v-adpr: -0.0918 0.0376 0.1652 0.1285 0.0380 0.0393 --------------------------------------------------------------- Specif. indices Q10 median Q90 CI80 mean full ==== MODEL 3 (Does not do well in terms of BIC) ===== Specif. indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- Param ter: 0.2909 0.3919 0.5669 0.1380 0.4157 0.3880 Param v1 easy: 0.4580 0.5519 0.6352 0.0886 0.5494 0.5503 Param v2 hard: 0.5667 0.6685 0.7693 0.1013 0.6682 0.6685 --------------------------------------------------------------- Avg v: 0.5049 0.5974 0.6799 0.0875 0.5942 0.5950 Gain=v-adpr: -0.1428 -0.0062 0.1198 0.1313 -0.0085 -0.0085 --------------------------------------------------------------- Specif. indices Q10 median Q90 CI80 mean full ==== MODEL 4 ===== Specif. indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- Param ter: 0.2391 0.3160 0.4165 0.0887 0.3225 0.3174 Param st: 0.1353 0.2260 0.3280 0.0963 0.2287 0.2316 Param v1 easy: 0.4764 0.5653 0.6544 0.0890 0.5627 0.5642 Param v2 hard: 0.5584 0.6620 0.7690 0.1053 0.6641 0.6636 --------------------------------------------------------------- Avg v: 0.5146 0.6045 0.6948 0.0901 0.6020 0.6029 Gain=v-adpr: -0.1357 -0.0057 0.1466 0.1412 -0.0007 -0.0005 t1=Ter-st/2: 0.2711 0.4778 1.1446 0.4367 0.6239 0.4890 t2=Ter+st/2: 0.2042 0.2803 0.3602 0.0780 0.2825 0.2831 --------------------------------------------------------------- Specif. indices Q10 median Q90 CI80 mean full
Learning-index summary table
fprintf('Learn indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('All dprim: %s\n', SI_stats_helper(LI_adpr_b)) ; fprintf('Easy dprim: %s\n', SI_stats_helper(LI_edpr_b)) ; fprintf('Hard dprim: %s\n', SI_stats_helper(LI_hdpr_b)) ; fprintf('RT_all: %s\n', SI_stats_helper(LI_RT_all_b)) ; fprintf('RT_correct: %s\n', SI_stats_helper(LI_RT_corr_b)) ; fprintf('RT_all std: %s\n', SI_stats_helper(LI_RTsd_all_b)) ; fprintf('RT_corr std: %s\n', SI_stats_helper(LI_RTsd_corr_b)) ; fprintf('---------------------------------------------------------------\n') fprintf('Param a: %s\n', SI_stats_helper(LI_a_b)) ; fprintf('Param ter: %s\n', SI_stats_helper(LI_ter_b)) ; fprintf('Param eta: %s\n', SI_stats_helper(LI_eta_b)) ; fprintf('Param sz: %s\n', SI_stats_helper(LI_sz_b)) ; fprintf('Param st: %s\n', SI_stats_helper(LI_st_b)) ; fprintf('Param v1 easy: %s\n', SI_stats_helper(LI_v1_b)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(LI_v2_b)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(LI_v_avg_b)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(LI_gain_b)) ; fprintf('t1=Ter-st/2: %s\n', SI_stats_helper(LI_t1_b)) ; fprintf('t2=Ter+st/2: %s\n', SI_stats_helper(LI_t2_b)) ; fprintf('---------------------------------------------------------------\n') fprintf('Learn indices Q10 median Q90 CI80 mean full\n') % Model 2 fprintf('\n\n==== MODEL 2 (Does not do well in terms of BIC) =====\n') ; fprintf('Learn indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('Param v1 easy: %s\n', SI_stats_helper(LI_v1_b_2)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(LI_v2_b_2)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(LI_v_avg_b_2)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(LI_gain_b_2)) ; fprintf('---------------------------------------------------------------\n') fprintf('Learn indices Q10 median Q90 CI80 mean full\n') % Model 3 fprintf('\n\n==== MODEL 3 (Does not do well in terms of BIC) =====\n') ; fprintf('Learn indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('Param ter: %s\n', SI_stats_helper(LI_ter_b_3)) ; fprintf('Param v1 easy: %s\n', SI_stats_helper(LI_v1_b_3)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(LI_v2_b_3)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(LI_v_avg_b_3)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(LI_gain_b_3)) ; fprintf('---------------------------------------------------------------\n') fprintf('Learn indices Q10 median Q90 CI80 mean full\n') % Model 4 fprintf('\n\n==== MODEL 4 =====\n') ; fprintf('Learn indices Q10 median Q90 CI80 mean full\n') fprintf('---------------------------------------------------------------\n') fprintf('Param ter: %s\n', SI_stats_helper(LI_ter_b_4)) ; fprintf('Param st: %s\n', SI_stats_helper(LI_st_b_4)) ; fprintf('Param v1 easy: %s\n', SI_stats_helper(LI_v1_b_4)) ; fprintf('Param v2 hard: %s\n', SI_stats_helper(LI_v2_b_4)) ; fprintf('---------------------------------------------------------------\n') fprintf('Avg v: %s\n', SI_stats_helper(LI_v_avg_b_4)) ; fprintf('Gain=v-adpr: %s\n', SI_stats_helper(LI_gain_b_4)) ; fprintf('t1=Ter-st/2: %s\n', SI_stats_helper(LI_t1_b_4)) ; fprintf('t2=Ter+st/2: %s\n', SI_stats_helper(LI_t2_b_4)) ; fprintf('---------------------------------------------------------------\n') fprintf('Learn indices Q10 median Q90 CI80 mean full\n')
Learn indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- All dprim: 0.4710 0.5438 0.6343 0.0817 0.5486 0.5478 Easy dprim: 0.4738 0.5531 0.6404 0.0833 0.5550 0.5554 Hard dprim: 0.5566 0.6502 0.7628 0.1031 0.6549 0.6525 RT_all: -0.3044 -0.2724 -0.2404 0.0320 -0.2723 -0.2734 RT_correct: -0.3027 -0.2724 -0.2407 0.0310 -0.2724 -0.2734 RT_all std: -0.1858 -0.1103 -0.0253 0.0802 -0.1095 -0.1072 RT_corr std: -0.1968 -0.1259 -0.0451 0.0758 -0.1251 -0.1236 --------------------------------------------------------------- Param a: -0.1617 -0.0918 -0.0217 0.0700 -0.0911 -0.0909 Param ter: -0.2443 -0.2043 -0.1591 0.0426 -0.2026 -0.2040 Param eta: 0.1732 0.3714 0.5964 0.2116 0.3811 0.3710 Param sz: -0.2419 -0.0233 0.2696 0.2558 0.0031 -0.0326 Param st: -0.6163 -0.5601 -0.4980 0.0591 -0.5573 -0.5582 Param v1 easy: 0.7388 0.9571 1.1988 0.2300 0.9607 0.9573 Param v2 hard: 0.8082 1.0478 1.3083 0.2500 1.0539 1.0469 --------------------------------------------------------------- Avg v: 0.7690 0.9880 1.2335 0.2323 0.9947 0.9902 Gain=v-adpr: 0.2404 0.4444 0.6523 0.2059 0.4462 0.4424 t1=Ter-st/2: -0.1477 -0.0664 0.0206 0.0841 -0.0640 -0.0682 t2=Ter+st/2: -0.3099 -0.2816 -0.2486 0.0306 -0.2802 -0.2809 --------------------------------------------------------------- Learn indices Q10 median Q90 CI80 mean full ==== MODEL 2 (Does not do well in terms of BIC) ===== Learn indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- Param v1 easy: 0.8968 1.0275 1.1642 0.1337 1.0295 1.0282 Param v2 hard: 0.7750 0.9085 1.0444 0.1347 0.9111 0.9050 --------------------------------------------------------------- Avg v: 0.8581 0.9823 1.1081 0.1250 0.9842 0.9814 Gain=v-adpr: 0.2883 0.4365 0.5747 0.1432 0.4356 0.4336 --------------------------------------------------------------- Learn indices Q10 median Q90 CI80 mean full ==== MODEL 3 (Does not do well in terms of BIC) ===== Learn indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- Param ter: -0.2513 -0.2023 -0.1474 0.0519 -0.2007 -0.2040 Param v1 easy: 0.6618 0.7739 0.8911 0.1146 0.7760 0.7731 Param v2 hard: 0.6843 0.7925 0.9018 0.1088 0.7933 0.7936 --------------------------------------------------------------- Avg v: 0.6750 0.7820 0.8917 0.1084 0.7822 0.7807 Gain=v-adpr: 0.1034 0.2306 0.3694 0.1330 0.2336 0.2329 --------------------------------------------------------------- Learn indices Q10 median Q90 CI80 mean full ==== MODEL 4 ===== Learn indices Q10 median Q90 CI80 mean full --------------------------------------------------------------- Param ter: -0.2984 -0.2604 -0.2159 0.0412 -0.2587 -0.2588 Param st: -0.6748 -0.6200 -0.5474 0.0637 -0.6156 -0.6169 Param v1 easy: 0.5797 0.6786 0.7854 0.1028 0.6805 0.6760 Param v2 hard: 0.6352 0.7455 0.8647 0.1147 0.7458 0.7411 --------------------------------------------------------------- Avg v: 0.6028 0.7015 0.8065 0.1019 0.7042 0.6999 Gain=v-adpr: 0.0226 0.1549 0.2902 0.1338 0.1556 0.1521 t1=Ter-st/2: -0.1914 -0.1201 -0.0437 0.0738 -0.1177 -0.1198 t2=Ter+st/2: -0.3717 -0.3380 -0.3023 0.0347 -0.3373 -0.3370 --------------------------------------------------------------- Learn indices Q10 median Q90 CI80 mean full
Bootstrap group-level Z-tests about specificity indices, accuracy
This is pre-October 2010, still works because it does not depend on explicit time variables.
% z-test for SI_dpr_diff_b = SI_hdpr_b - SI_edpr_b to determine whether % the easy and difficult d' have statistically different SIs SI_dpr_diff_b = SI_hdpr_b - SI_edpr_b ; % [N_samples, 1] SI_dpr_diff_stats.mean = mean(SI_dpr_diff_b) ; SI_dpr_diff_stats.std = std(SI_dpr_diff_b) ; SI_dpr_diff_stats.z = SI_dpr_diff_stats.mean ./ SI_dpr_diff_stats.std ; SI_dpr_diff_stats.onetailed_p = 1 - normcdf(SI_dpr_diff_stats.z) % z-test for SI_v_diff_b = SI_v2_b - SI_v1_b to determine whether % the easy and difficult drift rates have statistically different SIs SI_v_diff_b = SI_v2_b - SI_v1_b ; % [N_samples, 1] SI_v_diff_stats.mean = mean(SI_v_diff_b) ; SI_v_diff_stats.std = std(SI_v_diff_b) ; SI_v_diff_stats.z = SI_v_diff_stats.mean ./ SI_v_diff_stats.std ; SI_v_diff_stats.onetailed_p = 1 - normcdf(SI_v_diff_stats.z) % z-test for gain_SI := v_avg_SI - adpr_SI SI_gain_stats.mean = mean(SI_gain_b) ; SI_gain_stats.std = std(SI_gain_b) ; SI_gain_stats.z = SI_gain_stats.mean ./ SI_gain_stats.std ; SI_gain_stats.onetailed_p = 1 - normcdf(SI_gain_stats.z) % Conclusion: The gain in specificity does not reach statistical signif.
SI_dpr_diff_stats =
mean: 0.1109
std: 0.0741
z: 1.4962
onetailed_p: 0.0673
SI_v_diff_stats =
mean: 0.0572
std: 0.0386
z: 1.4811
onetailed_p: 0.0693
SI_gain_stats =
mean: 0.0850
std: 0.0930
z: 0.9141
onetailed_p: 0.1803
Bootstrap group-level Z-tests about Learning indices, accuracy
This is pre-October 2010, still works because it does not depend on explicit time variables.
% z-test for LI_dpr_diff_b = LI_hdpr_b - LI_edpr_b to determine whether % the easy and difficult d' have statistically different SIs LI_dpr_diff_b = LI_hdpr_b - LI_edpr_b ; % [N_samples, 1] LI_dpr_diff_stats.mean = mean(LI_dpr_diff_b) ; LI_dpr_diff_stats.std = std(LI_dpr_diff_b) ; LI_dpr_diff_stats.z = LI_dpr_diff_stats.mean ./ LI_dpr_diff_stats.std ; LI_dpr_diff_stats.onetailed_p = 1 - normcdf(LI_dpr_diff_stats.z) % z-test for LI_v_diff_b = LI_v2_b - LI_v1_b to determine whether % the easy and difficult drift rates have statistically different SIs LI_v_diff_b = LI_v2_b - LI_v1_b ; % [N_samples, 1] LI_v_diff_stats.mean = mean(LI_v_diff_b) ; LI_v_diff_stats.std = std(LI_v_diff_b) ; LI_v_diff_stats.z = LI_v_diff_stats.mean ./ LI_v_diff_stats.std ; LI_v_diff_stats.onetailed_p = 1 - normcdf(LI_v_diff_stats.z) % z-test for LI_gain_b := LI_v_avg_b - LI_adpr_b LI_gain_b = LI_v_avg_b - LI_adpr_b ; LI_gain_stats.mean = mean(LI_gain_b) ; LI_gain_stats.std = std(LI_gain_b) ; LI_gain_stats.z = LI_gain_stats.mean ./ LI_gain_stats.std ; LI_gain_stats.onetailed_p = 1 - normcdf(LI_gain_stats.z)
LI_dpr_diff_stats =
mean: 0.0999
std: 0.0495
z: 2.0169
onetailed_p: 0.0219
LI_v_diff_stats =
mean: 0.0931
std: 0.0607
z: 1.5343
onetailed_p: 0.0625
LI_gain_stats =
mean: 0.4462
std: 0.1599
z: 2.7898
onetailed_p: 0.0026
Bootstrap group-level Z-tests comparing the indices for Ter and MeanRT
Added by Alex, 2010-11-17.
% z-test for SI_gain_Ter_mRT := ter_SI - RT_all_SI SI_gain_Ter_mRT_stats.gain = [B.ter_SI]' - [B.RT_all_SI]' ; % [N_samples,1] SI_gain_Ter_mRT_stats.mean = mean(SI_gain_Ter_mRT_stats.gain) ; SI_gain_Ter_mRT_stats.std = std(SI_gain_Ter_mRT_stats.gain) ; SI_gain_Ter_mRT_stats.z = SI_gain_Ter_mRT_stats.mean ./ SI_gain_Ter_mRT_stats.std ; SI_gain_Ter_mRT_stats.onetailed_p = normcdf(SI_gain_Ter_mRT_stats.z) % Conclusion: The drop of the specificity index is marginally significant % z-test for LI_gain_Ter_mRT := ter_LI - RT_all_LI LI_gain_Ter_mRT_stats.gain = [B.ter_LI]' - [B.RT_all_LI]' ; % [N_samples,1] LI_gain_Ter_mRT_stats.mean = mean(LI_gain_Ter_mRT_stats.gain) ; LI_gain_Ter_mRT_stats.std = std(LI_gain_Ter_mRT_stats.gain) ; LI_gain_Ter_mRT_stats.z = LI_gain_Ter_mRT_stats.mean ./ LI_gain_Ter_mRT_stats.std ; LI_gain_Ter_mRT_stats.onetailed_p = 1 - normcdf(LI_gain_Ter_mRT_stats.z) % Conclusion: The drop of the learning index is statisctically significant
SI_gain_Ter_mRT_stats =
gain: [1000x1 double]
mean: -0.1463
std: 0.0680
z: -2.1536
onetailed_p: 0.0156
LI_gain_Ter_mRT_stats =
gain: [1000x1 double]
mean: 0.0697
std: 0.0279
z: 2.4954
onetailed_p: 0.0063
Individual-level Learning indices, accuracy and drift rates
This is pre-October 2010, updated for 11 periods.
LI_adpr_ind = learn_idx(dp11') ; % [N_sbj, 1] LI_edpr_ind = learn_idx(edp11') ; % [N_sbj, 1] LI_hdpr_ind = learn_idx(hdp11') ; % [N_sbj, 1] LI_dpr_diff_ind = LI_hdpr_ind - LI_edpr_ind ; describe([LI_adpr_ind LI_edpr_ind LI_hdpr_ind LI_dpr_diff_ind], ... {'LI_adpr_ind' 'LI_edpr_ind' 'LI_hdpr_ind' 'LI_dpr_diff_ind, hard-easy'}) % 80% confidence intervals CI80_multiplier = norminv(.90)/sqrt(N_sbj) CI80_multiplier.*std([LI_adpr_ind LI_edpr_ind LI_hdpr_ind LI_dpr_diff_ind]) LI_v_avg_ind = learn_idx((sbj_means(:,v1)+sbj_means(:,v2))./2) ; LI_v1_ind = learn_idx(sbj_means(:,v1)) ; % [N_sbj, 1] LI_v2_ind = learn_idx(sbj_means(:,v2)) ; % [N_sbj, 1] LI_v_diff_ind = LI_v2_ind - LI_v1_ind ; describe([LI_v_avg_ind LI_v1_ind LI_v2_ind LI_v_diff_ind], ... {'LI_v_avg_ind' 'LI_v1_ind, easy' 'LI_v2_ind, hard' 'LI_v_diff_ind, hard-easy'}) CI80_multiplier.*std([LI_v_avg_ind LI_v1_ind LI_v2_ind LI_v_diff_ind])
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
0.606 0.419 0.09 0.26 0.53 0.85 1.76 LI_adpr_ind
0.620 0.453 -0.14 0.36 0.47 0.83 2.07 LI_edpr_ind
0.721 0.499 0.07 0.35 0.62 0.94 1.88 LI_hdpr_ind
0.102 0.333 -0.67 -0.06 0.11 0.24 0.83 LI_dpr_diff_ind, hard-easy
------------------------------------------------------------
0.512 0.426 -0.16 0.23 0.43 0.71 1.64
CI80_multiplier =
0.2466
ans =
0.1034 0.1118 0.1231 0.0822
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
1.355 1.124 -0.41 0.53 1.09 1.97 4.33 LI_v_avg_ind
1.312 1.071 -0.45 0.48 1.18 1.85 3.94 LI_v1_ind, easy
1.460 1.265 -0.32 0.58 1.28 2.17 5.08 LI_v2_ind, hard
0.147 0.440 -0.89 -0.13 0.05 0.30 1.16 LI_v_diff_ind, hard-easy
------------------------------------------------------------
1.069 0.975 -0.52 0.37 0.90 1.57 3.63
ans =
0.2771 0.2642 0.3119 0.1086
Paired-sample t-tests of LIs, individual-subject data
This is pre-October 2010, still works because it does not depend on explicit time variables.
[k,p,k,LI_dpr_diff_stats] = ttest(LI_hdpr_ind-LI_edpr_ind) ; LI_dpr_diff_stats.mean = mean(LI_hdpr_ind-LI_edpr_ind) ; LI_dpr_diff_stats.onetailed_p = p/2 [k,p,k,LI_v_diff_stats] = ttest(LI_v2_ind-LI_v1_ind) ; LI_v_diff_stats.mean = mean(LI_v2_ind-LI_v1_ind) ; LI_v_diff_stats.onetailed_p = p/2
LI_dpr_diff_stats =
tstat: 1.5822
df: 26
sd: 0.3333
mean: 0.1015
onetailed_p: 0.0628
LI_v_diff_stats =
tstat: 1.7398
df: 26
sd: 0.4405
mean: 0.1475
onetailed_p: 0.0469
Proportionality of easy and hard d'
The ratio of the two stimulus deltas is 7/4 = 1.75 This is pre-October 2010, updated for 11 periods.
grand_mean_edp11 = mean(edp11') %#ok<*UDIM> % mean([N_sbj,11])=[1,11] grand_mean_hdp11 = mean(hdp11') grand_mean_dp11 = mean(dp11') ratio_edp11_hdp11_1x11 = grand_mean_edp11./grand_mean_hdp11 describe(ratio_edp11_hdp11_1x11,'edp11/hdp11, averaged across subjects') norminv(.90)*std(ratio_edp11_hdp11_1x11) ratio_edp11_hdp11_27x1 = (mean(edp11)./mean(hdp11))' ; describe(ratio_edp11_hdp11_27x1,'edp11/hdp11, averaged across periods') norminv(.90)*std(ratio_edp11_hdp11_27x1)
grand_mean_edp11 =
1.9742 2.4024 2.7152 2.8128 2.8620 3.0312 3.0682 3.0706 2.6388 2.4638 2.8112
grand_mean_hdp11 =
1.1553 1.4729 1.5972 1.7069 1.7923 1.8890 1.8887 1.9091 1.6944 1.4125 1.7599
grand_mean_dp11 =
1.5379 1.8925 2.0733 2.1644 2.2251 2.3292 2.3588 2.3805 2.1424 1.8721 2.1925
ratio_edp11_hdp11_1x11 =
1.7088 1.6310 1.7001 1.6479 1.5969 1.6046 1.6245 1.6084 1.5574 1.7442 1.5974
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
1.638 0.057 1.56 1.60 1.62 1.69 1.74 edp11/hdp11, averaged across subjects
ans =
0.0728
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
1.653 0.101 1.50 1.56 1.67 1.70 1.87 edp11/hdp11, averaged across periods
ans =
0.1292
Within-sbj ANOVA for the boundary separation parameter a
See Figure 14.1 in Howell (1992), "Statistical Methods for Psychology"
Verify that the boundary separation doesn't change significantly.
The first bootstrap "group" in B(1) contains data from all 27 subjects
%a27x11=sbj_means(:,a) ; % [N_sbj x N_periods], dependent var sbj=repmat((1:N_sbj)',1,N_periods_total) ; % IV period=repmat(1:N_periods_total,N_sbj,1) ; % IV [SS, df, MS, lbl] = anova(B(1).a(:),[sbj(:) period(:)],'SP') ; a_ANOVA.labels = lbl' ; a_ANOVA.SS = SS' ; a_ANOVA.MS = MS' ; a_ANOVA.df = df' ; a_ANOVA.F = MS(2)/MS(3) ; % MS('P')/MS('SxP') a_ANOVA.p = 1-fcdf(a_ANOVA.F,df(2),df(3)) clear SS df MS lbl
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 0.145 49.56 26 0.0056
2 P 0.008 2.72 10 0.0008
3 SP 0.139 47.72 260 0.0005
4 err -0.000 -0.00 0 0.0000
5 Totl 0.292 100.00 296 0.0010
------------------------------------------------------
a_ANOVA =
labels: [4x5 char]
SS: [0.1447 0.0079 0.1393 -1.7764e-15 0.2920]
MS: [0.0056 7.9286e-04 5.3592e-04 0 9.8647e-04]
df: [26 10 260 0 296]
F: 1.4794
p: 0.1470
Define linear and quadratic trend coefficients
2010-10-22 Doing a linear trend analysis is better than doing a simple pairwise comparison between points 1 and 8 because a "linear trend analysis gives progressively more weight to the treatment conditions located father away from the center of the independent variable. ... [pairwise comparisons] only incidentally represent the full extent of the linear trend."
The following equations are based on pg 65-92 of Keppel & Wickens "Design and Analysis: A Researcher's Handbook." (2004)
% Linear contrast coefficients: % Our independent variables (periods 1-8) are equidistant from one another... % The simplest set then is to use the independent variables themselves: lin_coef = 1:N_periods_train ; % They must satisfy the requirement that they sum to zero. This can be achieved % by subtracting out the mean from each lin_coef = lin_coef - mean(lin_coef) ; % Also, it is convenient to arrange that sum(lin_coef.^2)==1 lin_coef = lin_coef ./ sqrt(sum(lin_coef.^2)) % Sanity checks zerop = @(x) (abs(x)<1e-6) ; % TRUE if x equals zero within rounding error assert(zerop(sum(lin_coef))) ; % zero mean assert(zerop(sum(lin_coef.^2)-1)) ; % unit length % To test whether we need to account for a quadratic element, we must have % coefficients that are orthogonal to one another. These are sensitive to curvature % but not to any linear trend present. I (NVH) have chosen coef out of a table in % Keppel & Wickens "Design and Analysis: A Researcher's Handbook." (2004) Appendix A.3 % The sign is irrelevant because the SS does not depend on the sign. quad_coef = [7 1 -3 -5 -5 -3 1 7]; % These are orthogonal and sum to zero % Again, it is convenient to arrange that sum(quad_coef.^2)==1 quad_coef = quad_coef ./ sqrt(sum(quad_coef.^2)) % Sanity checks assert(zerop(sum(quad_coef))) ; % zero mean assert(zerop(sum(quad_coef.^2)-1)) ; % unit length % Verify orthogonality -- the dot product should be zero (within rounding error) assert(zerop(lin_coef*quad_coef')) ; clf plot(lin_coef,lin_coef,'b.-',lin_coef,quad_coef,'r.-') ;
lin_coef =
-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401
quad_coef =
0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401
Linear trend analysis of boundary separation parameter a
Added by Nick 2010-10-20. All diffusion parameters come from the saturated model (aka MODEL_1). Revised by Alex 2010-20-22.
% We are only looking at the training portion (periods 1-8) sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test a27x8=B(1).a(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(a27x8(:),[sbj(:) period(:)],'SP') ; trend_a_ANOVA.labels = lbl' ; trend_a_ANOVA.means = mean(a27x8) ; trend_a_ANOVA.SS = SS' ; trend_a_ANOVA.MS = MS' ; trend_a_ANOVA.df = df' ; trend_a_ANOVA.MS_err = MS(3) ; trend_a_ANOVA.df_err = df(3) ; trend_a_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_a_ANOVA.p_omnibus = 1-fcdf(trend_a_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_a_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_a_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_a_ANOVA.linear_contrast_coefs = lin_coef ; trend_a_ANOVA.psi_hat_linear = trend_a_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_a_ANOVA.SS_linear = N_sbj * trend_a_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_a_ANOVA.F_linear = trend_a_ANOVA.SS_linear / trend_a_ANOVA.MS_err ; trend_a_ANOVA.p_linear = 1-fcdf(trend_a_ANOVA.F_linear,1,trend_a_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_a_ANOVA.F_linear-1) ; trend_a_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_a_ANOVA.SS_failure_linear = trend_a_ANOVA.SS(2) - trend_a_ANOVA.SS_linear ; trend_a_ANOVA.df_failure_linear = trend_a_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_a_ANOVA.MS_failure_linear = trend_a_ANOVA.SS_failure_linear / trend_a_ANOVA.df_failure_linear ; trend_a_ANOVA.F_failure_linear = trend_a_ANOVA.MS_failure_linear / trend_a_ANOVA.MS_err ; trend_a_ANOVA.p_failure_linear = 1-fcdf(trend_a_ANOVA.F_failure_linear,... trend_a_ANOVA.df_failure_linear, trend_a_ANOVA.df_err) % % Where F_{.05} = % fprintf('F{.05} critical = %f, df=(%d,%d)\n', ... % finv(.95, trend_a_ANOVA.df_failure_linear, trend_a_ANOVA.df_err),... % trend_a_ANOVA.df_failure_linear, trend_a_ANOVA.df_err) ; % % Where F_{.10} = % fprintf('F{.10} critical = %f, df=(%d,%d)\n', ... % finv(.90, trend_a_ANOVA.df_failure_linear, trend_a_ANOVA.df_err),... % trend_a_ANOVA.df_failure_linear, trend_a_ANOVA.df_err) ; % If F_failure_linear is not significant, then there is reason to doubt that % a quadratic trend is necessary. However, one can check for this anyways (see below) clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 0.113 49.91 26 0.0044
2 P 0.007 3.04 7 0.0010
3 SP 0.107 47.05 182 0.0006
4 err 0.000 0.00 0 0.0000
5 Totl 0.227 100.00 215 0.0011
------------------------------------------------------
trend_a_ANOVA =
labels: [4x5 char]
means: [0.1361 0.1415 0.1418 0.1316 0.1346 0.1292 0.1331 0.1238]
SS: [0.1133 0.0069 0.1068 1.7764e-15 0.2269]
MS: [0.0044 9.8560e-04 5.8656e-04 0 0.0011]
df: [26 7 182 0 215]
MS_err: 5.8656e-04
df_err: 182
F_omnibus: 1.6803
p_omnibus: 0.1163
omega2_omnibus: 0.0185
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -0.0126
SS_linear: 0.0043
F_linear: 7.3228
p_linear: 0.0075
omega2_linear: 0.1048
SS_failure_linear: 0.0026
df_failure_linear: 6
MS_failure_linear: 4.3398e-04
F_failure_linear: 0.7399
p_failure_linear: 0.6182
Quadratic trend analysis of boundary separation parameter a
Added by Nick 2010-10-20. All diffusion parameters come from the saturated model (aka MODEL_1). Revised by Alex 2010-20-22.
% Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_a_ANOVA.quad_contrast_coefs = quad_coef ; trend_a_ANOVA.psi_hat_quad = trend_a_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_a_ANOVA.SS_quad = N_sbj * trend_a_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_a_ANOVA.F_quad = trend_a_ANOVA.SS_quad / trend_a_ANOVA.MS_err ; trend_a_ANOVA.p_quad = 1-fcdf(trend_a_ANOVA.F_quad,1,trend_a_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_a_ANOVA.F_quad-1) ; trend_a_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04
trend_a_ANOVA =
labels: [4x5 char]
means: [0.1361 0.1415 0.1418 0.1316 0.1346 0.1292 0.1331 0.1238]
SS: [0.1133 0.0069 0.1068 1.7764e-15 0.2269]
MS: [0.0044 9.8560e-04 5.8656e-04 0 0.0011]
df: [26 7 182 0 215]
MS_err: 5.8656e-04
df_err: 182
F_omnibus: 1.6803
p_omnibus: 0.1163
omega2_omnibus: 0.0185
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -0.0126
SS_linear: 0.0043
F_linear: 7.3228
p_linear: 0.0075
omega2_linear: 0.1048
SS_failure_linear: 0.0026
df_failure_linear: 6
MS_failure_linear: 4.3398e-04
F_failure_linear: 0.7399
p_failure_linear: 0.6182
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0038
SS_quad: 3.9822e-04
F_quad: 0.6789
p_quad: 0.4110
omega2_quad: 0
Linear (and quadratic) trend analysis of drift-stdev parameter eta
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test eta27x8=B(1).eta(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(eta27x8(:),[sbj(:) period(:)],'SP') ; trend_eta_ANOVA.labels = lbl' ; trend_eta_ANOVA.means = mean(eta27x8) ; trend_eta_ANOVA.SS = SS' ; trend_eta_ANOVA.MS = MS' ; trend_eta_ANOVA.df = df' ; trend_eta_ANOVA.MS_err = MS(3) ; trend_eta_ANOVA.df_err = df(3) ; trend_eta_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_eta_ANOVA.p_omnibus = 1-fcdf(trend_eta_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_eta_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_eta_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_eta_ANOVA.linear_contrast_coefs = lin_coef ; trend_eta_ANOVA.psi_hat_linear = trend_eta_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_eta_ANOVA.SS_linear = N_sbj * trend_eta_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_eta_ANOVA.F_linear = trend_eta_ANOVA.SS_linear / trend_eta_ANOVA.MS_err ; trend_eta_ANOVA.p_linear = 1-fcdf(trend_eta_ANOVA.F_linear,1,trend_eta_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_eta_ANOVA.F_linear-1) ; trend_eta_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_eta_ANOVA.SS_failure_linear = trend_eta_ANOVA.SS(2) - trend_eta_ANOVA.SS_linear ; trend_eta_ANOVA.df_failure_linear = trend_eta_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_eta_ANOVA.MS_failure_linear = trend_eta_ANOVA.SS_failure_linear / trend_eta_ANOVA.df_failure_linear ; trend_eta_ANOVA.F_failure_linear = trend_eta_ANOVA.MS_failure_linear / trend_eta_ANOVA.MS_err ; trend_eta_ANOVA.p_failure_linear = 1-fcdf(trend_eta_ANOVA.F_failure_linear,... trend_eta_ANOVA.df_failure_linear, trend_eta_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_eta_ANOVA.quad_contrast_coefs = quad_coef ; trend_eta_ANOVA.psi_hat_quad = trend_eta_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_eta_ANOVA.SS_quad = N_sbj * trend_eta_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_eta_ANOVA.F_quad = trend_eta_ANOVA.SS_quad / trend_eta_ANOVA.MS_err ; trend_eta_ANOVA.p_quad = 1-fcdf(trend_eta_ANOVA.F_quad,1,trend_eta_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_eta_ANOVA.F_quad-1) ; trend_eta_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 0.423 21.76 26 0.0163
2 P 0.109 5.59 7 0.0155
3 SP 1.411 72.65 182 0.0078
4 err -0.000 -0.00 0 0.0000
5 Totl 1.942 100.00 215 0.0090
------------------------------------------------------
trend_eta_ANOVA =
labels: [4x5 char]
means: [0.1974 0.2446 0.2556 0.2412 0.2765 0.2433 0.2500 0.2707]
SS: [0.4226 0.1086 1.4108 -7.1054e-15 1.9420]
MS: [0.0163 0.0155 0.0078 0 0.0090]
df: [26 7 182 0 215]
MS_err: 0.0078
df_err: 182
F_omnibus: 2.0015
p_omnibus: 0.0572
omega2_omnibus: 0.0271
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.0415
SS_linear: 0.0465
F_linear: 6.0054
p_linear: 0.0152
omega2_linear: 0.0848
SS_failure_linear: 0.0621
df_failure_linear: 6
MS_failure_linear: 0.0103
F_failure_linear: 1.3342
p_failure_linear: 0.2441
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0242
SS_quad: 0.0158
F_quad: 2.0380
p_quad: 0.1551
omega2_quad: 0.0189
Linear (and quadratic) trend analysis of starting-point range parameter sz
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test sz27x8=B(1).sz(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(sz27x8(:),[sbj(:) period(:)],'SP') ; trend_sz_ANOVA.labels = lbl' ; trend_sz_ANOVA.means = mean(sz27x8) ; trend_sz_ANOVA.SS = SS' ; trend_sz_ANOVA.MS = MS' ; trend_sz_ANOVA.df = df' ; trend_sz_ANOVA.MS_err = MS(3) ; trend_sz_ANOVA.df_err = df(3) ; trend_sz_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_sz_ANOVA.p_omnibus = 1-fcdf(trend_sz_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_sz_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_sz_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_sz_ANOVA.linear_contrast_coefs = lin_coef ; trend_sz_ANOVA.psi_hat_linear = trend_sz_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_sz_ANOVA.SS_linear = N_sbj * trend_sz_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_sz_ANOVA.F_linear = trend_sz_ANOVA.SS_linear / trend_sz_ANOVA.MS_err ; trend_sz_ANOVA.p_linear = 1-fcdf(trend_sz_ANOVA.F_linear,1,trend_sz_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_sz_ANOVA.F_linear-1) ; trend_sz_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_sz_ANOVA.SS_failure_linear = trend_sz_ANOVA.SS(2) - trend_sz_ANOVA.SS_linear ; trend_sz_ANOVA.df_failure_linear = trend_sz_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_sz_ANOVA.MS_failure_linear = trend_sz_ANOVA.SS_failure_linear / trend_sz_ANOVA.df_failure_linear ; trend_sz_ANOVA.F_failure_linear = trend_sz_ANOVA.MS_failure_linear / trend_sz_ANOVA.MS_err ; trend_sz_ANOVA.p_failure_linear = 1-fcdf(trend_sz_ANOVA.F_failure_linear,... trend_sz_ANOVA.df_failure_linear, trend_sz_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_sz_ANOVA.quad_contrast_coefs = quad_coef ; trend_sz_ANOVA.psi_hat_quad = trend_sz_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_sz_ANOVA.SS_quad = N_sbj * trend_sz_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_sz_ANOVA.F_quad = trend_sz_ANOVA.SS_quad / trend_sz_ANOVA.MS_err ; trend_sz_ANOVA.p_quad = 1-fcdf(trend_sz_ANOVA.F_quad,1,trend_sz_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_sz_ANOVA.F_quad-1) ; trend_sz_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 0.071 27.26 26 0.0027
2 P 0.008 3.21 7 0.0012
3 SP 0.180 69.53 182 0.0010
4 err 0.000 0.00 0 0.0000
5 Totl 0.259 100.00 215 0.0012
------------------------------------------------------
trend_sz_ANOVA =
labels: [4x5 char]
means: [0.0454 0.0458 0.0353 0.0407 0.0570 0.0521 0.0440 0.0440]
SS: [0.0705 0.0083 0.1799 4.4409e-16 0.2587]
MS: [0.0027 0.0012 9.8836e-04 0 0.0012]
df: [26 7 182 0 215]
MS_err: 9.8836e-04
df_err: 182
F_omnibus: 1.2007
p_omnibus: 0.3045
omega2_omnibus: 0.0055
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.0037
SS_linear: 3.6176e-04
F_linear: 0.3660
p_linear: 0.5459
omega2_linear: 0
SS_failure_linear: 0.0079
df_failure_linear: 6
MS_failure_linear: 0.0013
F_failure_linear: 1.3398
p_failure_linear: 0.2416
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0027
SS_quad: 1.9771e-04
F_quad: 0.2000
p_quad: 0.6552
omega2_quad: 0
Linear (and quadratic) trend analysis of easy drift rate v1
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test v1_27x8=B(1).v1(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(v1_27x8(:),[sbj(:) period(:)],'SP') ; trend_v1_ANOVA.labels = lbl' ; trend_v1_ANOVA.means = mean(v1_27x8) ; trend_v1_ANOVA.SS = SS' ; trend_v1_ANOVA.MS = MS' ; trend_v1_ANOVA.df = df' ; trend_v1_ANOVA.MS_err = MS(3) ; trend_v1_ANOVA.df_err = df(3) ; trend_v1_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_v1_ANOVA.p_omnibus = 1-fcdf(trend_v1_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_v1_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_v1_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_v1_ANOVA.linear_contrast_coefs = lin_coef ; trend_v1_ANOVA.psi_hat_linear = trend_v1_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_v1_ANOVA.SS_linear = N_sbj * trend_v1_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_v1_ANOVA.F_linear = trend_v1_ANOVA.SS_linear / trend_v1_ANOVA.MS_err ; trend_v1_ANOVA.p_linear = 1-fcdf(trend_v1_ANOVA.F_linear,1,trend_v1_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_v1_ANOVA.F_linear-1) ; trend_v1_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_v1_ANOVA.SS_failure_linear = trend_v1_ANOVA.SS(2) - trend_v1_ANOVA.SS_linear ; trend_v1_ANOVA.df_failure_linear = trend_v1_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_v1_ANOVA.MS_failure_linear = trend_v1_ANOVA.SS_failure_linear / trend_v1_ANOVA.df_failure_linear ; trend_v1_ANOVA.F_failure_linear = trend_v1_ANOVA.MS_failure_linear / trend_v1_ANOVA.MS_err ; trend_v1_ANOVA.p_failure_linear = 1-fcdf(trend_v1_ANOVA.F_failure_linear,... trend_v1_ANOVA.df_failure_linear, trend_v1_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_v1_ANOVA.quad_contrast_coefs = quad_coef ; trend_v1_ANOVA.psi_hat_quad = trend_v1_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_v1_ANOVA.SS_quad = N_sbj * trend_v1_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_v1_ANOVA.F_quad = trend_v1_ANOVA.SS_quad / trend_v1_ANOVA.MS_err ; trend_v1_ANOVA.p_quad = 1-fcdf(trend_v1_ANOVA.F_quad,1,trend_v1_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_v1_ANOVA.F_quad-1) ; trend_v1_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 2.222 42.49 26 0.0855
2 P 1.114 21.31 7 0.1592
3 SP 1.894 36.21 182 0.0104
4 err -0.000 -0.00 0 0.0000
5 Totl 5.230 100.00 215 0.0243
------------------------------------------------------
trend_v1_ANOVA =
labels: [4x5 char]
means: [0.2541 0.3517 0.3980 0.4133 0.4546 0.4477 0.4588 0.4973]
SS: [2.2219 1.1144 1.8935 -1.4211e-14 5.2298]
MS: [0.0855 0.1592 0.0104 0 0.0243]
df: [26 7 182 0 215]
MS_err: 0.0104
df_err: 182
F_omnibus: 15.3016
p_omnibus: 1.1102e-15
omega2_omnibus: 0.2843
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.1873
SS_linear: 0.9477
F_linear: 91.0905
p_linear: 0
omega2_linear: 0.6252
SS_failure_linear: 0.1667
df_failure_linear: 6
MS_failure_linear: 0.0278
F_failure_linear: 2.6701
p_failure_linear: 0.0166
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0622
SS_quad: 0.1046
F_quad: 10.0509
p_quad: 0.0018
omega2_quad: 0.1435
Linear (and quadratic) trend analysis of difficult drift rate v2
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test v2_27x8=B(1).v2(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(v2_27x8(:),[sbj(:) period(:)],'SP') ; trend_v2_ANOVA.labels = lbl' ; trend_v2_ANOVA.means = mean(v2_27x8) ; trend_v2_ANOVA.SS = SS' ; trend_v2_ANOVA.MS = MS' ; trend_v2_ANOVA.df = df' ; trend_v2_ANOVA.MS_err = MS(3) ; trend_v2_ANOVA.df_err = df(3) ; trend_v2_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_v2_ANOVA.p_omnibus = 1-fcdf(trend_v2_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_v2_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_v2_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_v2_ANOVA.linear_contrast_coefs = lin_coef ; trend_v2_ANOVA.psi_hat_linear = trend_v2_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_v2_ANOVA.SS_linear = N_sbj * trend_v2_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_v2_ANOVA.F_linear = trend_v2_ANOVA.SS_linear / trend_v2_ANOVA.MS_err ; trend_v2_ANOVA.p_linear = 1-fcdf(trend_v2_ANOVA.F_linear,1,trend_v2_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_v2_ANOVA.F_linear-1) ; trend_v2_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_v2_ANOVA.SS_failure_linear = trend_v2_ANOVA.SS(2) - trend_v2_ANOVA.SS_linear ; trend_v2_ANOVA.df_failure_linear = trend_v2_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_v2_ANOVA.MS_failure_linear = trend_v2_ANOVA.SS_failure_linear / trend_v2_ANOVA.df_failure_linear ; trend_v2_ANOVA.F_failure_linear = trend_v2_ANOVA.MS_failure_linear / trend_v2_ANOVA.MS_err ; trend_v2_ANOVA.p_failure_linear = 1-fcdf(trend_v2_ANOVA.F_failure_linear,... trend_v2_ANOVA.df_failure_linear, trend_v2_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_v2_ANOVA.quad_contrast_coefs = quad_coef ; trend_v2_ANOVA.psi_hat_quad = trend_v2_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_v2_ANOVA.SS_quad = N_sbj * trend_v2_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_v2_ANOVA.F_quad = trend_v2_ANOVA.SS_quad / trend_v2_ANOVA.MS_err ; trend_v2_ANOVA.p_quad = 1-fcdf(trend_v2_ANOVA.F_quad,1,trend_v2_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_v2_ANOVA.F_quad-1) ; trend_v2_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 1.022 45.18 26 0.0393
2 P 0.468 20.68 7 0.0668
3 SP 0.773 34.14 182 0.0042
4 err -0.000 -0.00 0 0.0000
5 Totl 2.262 100.00 215 0.0105
------------------------------------------------------
trend_v2_ANOVA =
labels: [4x5 char]
means: [0.1477 0.2118 0.2311 0.2497 0.2776 0.2769 0.2798 0.3023]
SS: [1.0221 0.4679 0.7725 -7.1054e-15 2.2625]
MS: [0.0393 0.0668 0.0042 0 0.0105]
df: [26 7 182 0 215]
MS_err: 0.0042
df_err: 182
F_omnibus: 15.7463
p_omnibus: 4.4409e-16
omega2_omnibus: 0.2906
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.1225
SS_linear: 0.4052
F_linear: 95.4601
p_linear: 0
omega2_linear: 0.6363
SS_failure_linear: 0.0627
df_failure_linear: 6
MS_failure_linear: 0.0104
F_failure_linear: 2.4607
p_failure_linear: 0.0260
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0400
SS_quad: 0.0432
F_quad: 10.1844
p_quad: 0.0017
omega2_quad: 0.1454
Linear (and quadratic) trend analysis of the average drift rate v_avg
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test v_avg_27x8=B(1).v_avg(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(v_avg_27x8(:),[sbj(:) period(:)],'SP') ; trend_v_avg_ANOVA.labels = lbl' ; trend_v_avg_ANOVA.means = mean(v_avg_27x8) ; trend_v_avg_ANOVA.SS = SS' ; trend_v_avg_ANOVA.MS = MS' ; trend_v_avg_ANOVA.df = df' ; trend_v_avg_ANOVA.MS_err = MS(3) ; trend_v_avg_ANOVA.df_err = df(3) ; trend_v_avg_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_v_avg_ANOVA.p_omnibus = 1-fcdf(trend_v_avg_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_v_avg_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_v_avg_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_v_avg_ANOVA.linear_contrast_coefs = lin_coef ; trend_v_avg_ANOVA.psi_hat_linear = trend_v_avg_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_v_avg_ANOVA.SS_linear = N_sbj * trend_v_avg_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_v_avg_ANOVA.F_linear = trend_v_avg_ANOVA.SS_linear / trend_v_avg_ANOVA.MS_err ; trend_v_avg_ANOVA.p_linear = 1-fcdf(trend_v_avg_ANOVA.F_linear,1,trend_v_avg_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_v_avg_ANOVA.F_linear-1) ; trend_v_avg_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_v_avg_ANOVA.SS_failure_linear = trend_v_avg_ANOVA.SS(2) - trend_v_avg_ANOVA.SS_linear ; trend_v_avg_ANOVA.df_failure_linear = trend_v_avg_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_v_avg_ANOVA.MS_failure_linear = trend_v_avg_ANOVA.SS_failure_linear / trend_v_avg_ANOVA.df_failure_linear ; trend_v_avg_ANOVA.F_failure_linear = trend_v_avg_ANOVA.MS_failure_linear / trend_v_avg_ANOVA.MS_err ; trend_v_avg_ANOVA.p_failure_linear = 1-fcdf(trend_v_avg_ANOVA.F_failure_linear,... trend_v_avg_ANOVA.df_failure_linear, trend_v_avg_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_v_avg_ANOVA.quad_contrast_coefs = quad_coef ; trend_v_avg_ANOVA.psi_hat_quad = trend_v_avg_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_v_avg_ANOVA.SS_quad = N_sbj * trend_v_avg_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_v_avg_ANOVA.F_quad = trend_v_avg_ANOVA.SS_quad / trend_v_avg_ANOVA.MS_err ; trend_v_avg_ANOVA.p_quad = 1-fcdf(trend_v_avg_ANOVA.F_quad,1,trend_v_avg_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_v_avg_ANOVA.F_quad-1) ; trend_v_avg_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 1.554 44.27 26 0.0598
2 P 0.755 21.52 7 0.1079
3 SP 1.201 34.21 182 0.0066
4 err -0.000 -0.00 0 0.0000
5 Totl 3.510 100.00 215 0.0163
------------------------------------------------------
trend_v_avg_ANOVA =
labels: [4x5 char]
means: [0.2009 0.2818 0.3145 0.3315 0.3661 0.3623 0.3693 0.3998]
SS: [1.5540 0.7555 1.2009 -3.5527e-15 3.5104]
MS: [0.0598 0.1079 0.0066 0 0.0163]
df: [26 7 182 0 215]
MS_err: 0.0066
df_err: 182
F_omnibus: 16.3552
p_omnibus: 1.1102e-16
omega2_omnibus: 0.2990
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.1549
SS_linear: 0.6481
F_linear: 98.2111
p_linear: 0
omega2_linear: 0.6429
SS_failure_linear: 0.1074
df_failure_linear: 6
MS_failure_linear: 0.0179
F_failure_linear: 2.7126
p_failure_linear: 0.0151
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0511
SS_quad: 0.0706
F_quad: 10.6941
p_quad: 0.0013
omega2_quad: 0.1522
Linear (and quadratic) trend analysis of the average dprime
Added by Alex 2010-11-10.
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test dpr_avg_27x8 = dp11(1:N_periods_train,:)' ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(dpr_avg_27x8(:),[sbj(:) period(:)],'SP') ; trend_dpr_avg_ANOVA.labels = lbl' ; trend_dpr_avg_ANOVA.means = mean(dpr_avg_27x8) ; trend_dpr_avg_ANOVA.SS = SS' ; trend_dpr_avg_ANOVA.MS = MS' ; trend_dpr_avg_ANOVA.df = df' ; trend_dpr_avg_ANOVA.MS_err = MS(3) ; trend_dpr_avg_ANOVA.df_err = df(3) ; trend_dpr_avg_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_dpr_avg_ANOVA.p_omnibus = 1-fcdf(trend_dpr_avg_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_dpr_avg_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_dpr_avg_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_dpr_avg_ANOVA.linear_contrast_coefs = lin_coef ; trend_dpr_avg_ANOVA.psi_hat_linear = trend_dpr_avg_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_dpr_avg_ANOVA.SS_linear = N_sbj * trend_dpr_avg_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_dpr_avg_ANOVA.F_linear = trend_dpr_avg_ANOVA.SS_linear / trend_dpr_avg_ANOVA.MS_err ; trend_dpr_avg_ANOVA.p_linear = 1-fcdf(trend_dpr_avg_ANOVA.F_linear,1,trend_dpr_avg_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_dpr_avg_ANOVA.F_linear-1) ; trend_dpr_avg_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_dpr_avg_ANOVA.SS_failure_linear = trend_dpr_avg_ANOVA.SS(2) - trend_dpr_avg_ANOVA.SS_linear ; trend_dpr_avg_ANOVA.df_failure_linear = trend_dpr_avg_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_dpr_avg_ANOVA.MS_failure_linear = trend_dpr_avg_ANOVA.SS_failure_linear / trend_dpr_avg_ANOVA.df_failure_linear ; trend_dpr_avg_ANOVA.F_failure_linear = trend_dpr_avg_ANOVA.MS_failure_linear / trend_dpr_avg_ANOVA.MS_err ; trend_dpr_avg_ANOVA.p_failure_linear = 1-fcdf(trend_dpr_avg_ANOVA.F_failure_linear,... trend_dpr_avg_ANOVA.df_failure_linear, trend_dpr_avg_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_dpr_avg_ANOVA.quad_contrast_coefs = quad_coef ; trend_dpr_avg_ANOVA.psi_hat_quad = trend_dpr_avg_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_dpr_avg_ANOVA.SS_quad = N_sbj * trend_dpr_avg_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_dpr_avg_ANOVA.F_quad = trend_dpr_avg_ANOVA.SS_quad / trend_dpr_avg_ANOVA.MS_err ; trend_dpr_avg_ANOVA.p_quad = 1-fcdf(trend_dpr_avg_ANOVA.F_quad,1,trend_dpr_avg_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_dpr_avg_ANOVA.F_quad-1) ; trend_dpr_avg_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 55.124 66.80 26 2.1202
2 P 15.509 18.79 7 2.2155
3 SP 11.894 14.41 182 0.0653
4 err 0.000 0.00 0 0.0000
5 Totl 82.526 100.00 215 0.3838
------------------------------------------------------
trend_dpr_avg_ANOVA =
labels: [4x5 char]
means: [1.5379 1.8925 2.0733 2.1644 2.2251 2.3292 2.3588 2.3805]
SS: [55.1242 15.5085 11.8937 2.2737e-13 82.5264]
MS: [2.1202 2.2155 0.0653 0 0.3838]
df: [26 7 182 0 215]
MS_err: 0.0653
df_err: 182
F_omnibus: 33.9022
p_omnibus: 0
omega2_omnibus: 0.4775
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.6988
SS_linear: 13.1849
F_linear: 201.7581
p_linear: 0
omega2_linear: 0.7880
SS_failure_linear: 2.3237
df_failure_linear: 6
MS_failure_linear: 0.3873
F_failure_linear: 5.9262
p_failure_linear: 1.1310e-05
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.2681
SS_quad: 1.9410
F_quad: 29.7019
p_quad: 1.6214e-07
omega2_quad: 0.3471
Linear (and quadratic) trend analysis of the easy dprime
Added by Alex 2010-11-10.
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test dpr_ez_27x8 = edp11(1:N_periods_train,:)' ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(dpr_ez_27x8(:),[sbj(:) period(:)],'SP') ; trend_dpr_ez_ANOVA.labels = lbl' ; trend_dpr_ez_ANOVA.means = mean(dpr_ez_27x8) ; trend_dpr_ez_ANOVA.SS = SS' ; trend_dpr_ez_ANOVA.MS = MS' ; trend_dpr_ez_ANOVA.df = df' ; trend_dpr_ez_ANOVA.MS_err = MS(3) ; trend_dpr_ez_ANOVA.df_err = df(3) ; trend_dpr_ez_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_dpr_ez_ANOVA.p_omnibus = 1-fcdf(trend_dpr_ez_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_dpr_ez_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_dpr_ez_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_dpr_ez_ANOVA.linear_contrast_coefs = lin_coef ; trend_dpr_ez_ANOVA.psi_hat_linear = trend_dpr_ez_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_dpr_ez_ANOVA.SS_linear = N_sbj * trend_dpr_ez_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_dpr_ez_ANOVA.F_linear = trend_dpr_ez_ANOVA.SS_linear / trend_dpr_ez_ANOVA.MS_err ; trend_dpr_ez_ANOVA.p_linear = 1-fcdf(trend_dpr_ez_ANOVA.F_linear,1,trend_dpr_ez_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_dpr_ez_ANOVA.F_linear-1) ; trend_dpr_ez_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_dpr_ez_ANOVA.SS_failure_linear = trend_dpr_ez_ANOVA.SS(2) - trend_dpr_ez_ANOVA.SS_linear ; trend_dpr_ez_ANOVA.df_failure_linear = trend_dpr_ez_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_dpr_ez_ANOVA.MS_failure_linear = trend_dpr_ez_ANOVA.SS_failure_linear / trend_dpr_ez_ANOVA.df_failure_linear ; trend_dpr_ez_ANOVA.F_failure_linear = trend_dpr_ez_ANOVA.MS_failure_linear / trend_dpr_ez_ANOVA.MS_err ; trend_dpr_ez_ANOVA.p_failure_linear = 1-fcdf(trend_dpr_ez_ANOVA.F_failure_linear,... trend_dpr_ez_ANOVA.df_failure_linear, trend_dpr_ez_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_dpr_ez_ANOVA.quad_contrast_coefs = quad_coef ; trend_dpr_ez_ANOVA.psi_hat_quad = trend_dpr_ez_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_dpr_ez_ANOVA.SS_quad = N_sbj * trend_dpr_ez_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_dpr_ez_ANOVA.F_quad = trend_dpr_ez_ANOVA.SS_quad / trend_dpr_ez_ANOVA.MS_err ; trend_dpr_ez_ANOVA.p_quad = 1-fcdf(trend_dpr_ez_ANOVA.F_quad,1,trend_dpr_ez_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_dpr_ez_ANOVA.F_quad-1) ; trend_dpr_ez_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 93.838 62.92 26 3.6092
2 P 27.622 18.52 7 3.9460
3 SP 27.674 18.56 182 0.1521
4 err 0.000 0.00 0 0.0000
5 Totl 149.134 100.00 215 0.6936
------------------------------------------------------
trend_dpr_ez_ANOVA =
labels: [4x5 char]
means: [1.9742 2.4024 2.7152 2.8128 2.8620 3.0312 3.0682 3.0706]
SS: [93.8382 27.6223 27.6738 6.8212e-13 149.1343]
MS: [3.6092 3.9460 0.1521 0 0.6936]
df: [26 7 182 0 215]
MS_err: 0.1521
df_err: 182
F_omnibus: 25.9516
p_omnibus: 0
omega2_omnibus: 0.4094
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.9259
SS_linear: 23.1470
F_linear: 152.2293
p_linear: 0
omega2_linear: 0.7369
SS_failure_linear: 4.4753
df_failure_linear: 6
MS_failure_linear: 0.7459
F_failure_linear: 4.9054
p_failure_linear: 1.1289e-04
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.3726
SS_quad: 3.7487
F_quad: 24.6539
p_quad: 1.5716e-06
omega2_quad: 0.3046
Linear (and quadratic) trend analysis of the hard dprime
Added by Alex 2010-11-10.
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test dpr_hd_27x8 = hdp11(1:N_periods_train,:)' ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(dpr_hd_27x8(:),[sbj(:) period(:)],'SP') ; trend_dpr_hd_ANOVA.labels = lbl' ; trend_dpr_hd_ANOVA.means = mean(dpr_hd_27x8) ; trend_dpr_hd_ANOVA.SS = SS' ; trend_dpr_hd_ANOVA.MS = MS' ; trend_dpr_hd_ANOVA.df = df' ; trend_dpr_hd_ANOVA.MS_err = MS(3) ; trend_dpr_hd_ANOVA.df_err = df(3) ; trend_dpr_hd_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_dpr_hd_ANOVA.p_omnibus = 1-fcdf(trend_dpr_hd_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_dpr_hd_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_dpr_hd_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_dpr_hd_ANOVA.linear_contrast_coefs = lin_coef ; trend_dpr_hd_ANOVA.psi_hat_linear = trend_dpr_hd_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_dpr_hd_ANOVA.SS_linear = N_sbj * trend_dpr_hd_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_dpr_hd_ANOVA.F_linear = trend_dpr_hd_ANOVA.SS_linear / trend_dpr_hd_ANOVA.MS_err ; trend_dpr_hd_ANOVA.p_linear = 1-fcdf(trend_dpr_hd_ANOVA.F_linear,1,trend_dpr_hd_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_dpr_hd_ANOVA.F_linear-1) ; trend_dpr_hd_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_dpr_hd_ANOVA.SS_failure_linear = trend_dpr_hd_ANOVA.SS(2) - trend_dpr_hd_ANOVA.SS_linear ; trend_dpr_hd_ANOVA.df_failure_linear = trend_dpr_hd_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_dpr_hd_ANOVA.MS_failure_linear = trend_dpr_hd_ANOVA.SS_failure_linear / trend_dpr_hd_ANOVA.df_failure_linear ; trend_dpr_hd_ANOVA.F_failure_linear = trend_dpr_hd_ANOVA.MS_failure_linear / trend_dpr_hd_ANOVA.MS_err ; trend_dpr_hd_ANOVA.p_failure_linear = 1-fcdf(trend_dpr_hd_ANOVA.F_failure_linear,... trend_dpr_hd_ANOVA.df_failure_linear, trend_dpr_hd_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_dpr_hd_ANOVA.quad_contrast_coefs = quad_coef ; trend_dpr_hd_ANOVA.psi_hat_quad = trend_dpr_hd_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_dpr_hd_ANOVA.SS_quad = N_sbj * trend_dpr_hd_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_dpr_hd_ANOVA.F_quad = trend_dpr_hd_ANOVA.SS_quad / trend_dpr_hd_ANOVA.MS_err ; trend_dpr_hd_ANOVA.p_quad = 1-fcdf(trend_dpr_hd_ANOVA.F_quad,1,trend_dpr_hd_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_dpr_hd_ANOVA.F_quad-1) ; trend_dpr_hd_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 45.352 64.32 26 1.7443
2 P 12.906 18.31 7 1.8438
3 SP 12.247 17.37 182 0.0673
4 err 0.000 0.00 0 0.0000
5 Totl 70.506 100.00 215 0.3279
------------------------------------------------------
trend_dpr_hd_ANOVA =
labels: [4x5 char]
means: [1.1553 1.4729 1.5972 1.7069 1.7923 1.8890 1.8887 1.9091]
SS: [45.3525 12.9063 12.2470 5.6843e-13 70.5057]
MS: [1.7443 1.8438 0.0673 0 0.3279]
df: [26 7 182 0 215]
MS_err: 0.0673
df_err: 182
F_omnibus: 27.3996
p_omnibus: 0
omega2_omnibus: 0.4231
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: 0.6416
SS_linear: 11.1156
F_linear: 165.1869
p_linear: 0
omega2_linear: 0.7525
SS_failure_linear: 1.7906
df_failure_linear: 6
MS_failure_linear: 0.2984
F_failure_linear: 4.4351
p_failure_linear: 3.2652e-04
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.2424
SS_quad: 1.5865
F_quad: 23.5764
p_quad: 2.5755e-06
omega2_quad: 0.2948
Linear (and quadratic) trend analysis of the mean raw RT (sRT_all)
Added by Alex 2010-11-10.
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test sRT_all_27x8 = sRT_all(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(sRT_all_27x8(:),[sbj(:) period(:)],'SP') ; trend_sRT_all_ANOVA.labels = lbl' ; trend_sRT_all_ANOVA.means = mean(sRT_all_27x8) ; trend_sRT_all_ANOVA.SS = SS' ; trend_sRT_all_ANOVA.MS = MS' ; trend_sRT_all_ANOVA.df = df' ; trend_sRT_all_ANOVA.MS_err = MS(3) ; trend_sRT_all_ANOVA.df_err = df(3) ; trend_sRT_all_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_sRT_all_ANOVA.p_omnibus = 1-fcdf(trend_sRT_all_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_sRT_all_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_sRT_all_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_sRT_all_ANOVA.linear_contrast_coefs = lin_coef ; trend_sRT_all_ANOVA.psi_hat_linear = trend_sRT_all_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_sRT_all_ANOVA.SS_linear = N_sbj * trend_sRT_all_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_sRT_all_ANOVA.F_linear = trend_sRT_all_ANOVA.SS_linear / trend_sRT_all_ANOVA.MS_err ; trend_sRT_all_ANOVA.p_linear = 1-fcdf(trend_sRT_all_ANOVA.F_linear,1,trend_sRT_all_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_sRT_all_ANOVA.F_linear-1) ; trend_sRT_all_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_sRT_all_ANOVA.SS_failure_linear = trend_sRT_all_ANOVA.SS(2) - trend_sRT_all_ANOVA.SS_linear ; trend_sRT_all_ANOVA.df_failure_linear = trend_sRT_all_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_sRT_all_ANOVA.MS_failure_linear = trend_sRT_all_ANOVA.SS_failure_linear / trend_sRT_all_ANOVA.df_failure_linear ; trend_sRT_all_ANOVA.F_failure_linear = trend_sRT_all_ANOVA.MS_failure_linear / trend_sRT_all_ANOVA.MS_err ; trend_sRT_all_ANOVA.p_failure_linear = 1-fcdf(trend_sRT_all_ANOVA.F_failure_linear,... trend_sRT_all_ANOVA.df_failure_linear, trend_sRT_all_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_sRT_all_ANOVA.quad_contrast_coefs = quad_coef ; trend_sRT_all_ANOVA.psi_hat_quad = trend_sRT_all_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_sRT_all_ANOVA.SS_quad = N_sbj * trend_sRT_all_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_sRT_all_ANOVA.F_quad = trend_sRT_all_ANOVA.SS_quad / trend_sRT_all_ANOVA.MS_err ; trend_sRT_all_ANOVA.p_quad = 1-fcdf(trend_sRT_all_ANOVA.F_quad,1,trend_sRT_all_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_sRT_all_ANOVA.F_quad-1) ; trend_sRT_all_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 1386926.318 46.73 26 53343.3199
2 P 956644.629 32.23 7 136663.5184
3 SP 624614.021 21.04 182 3431.9452
4 err 0.000 0.00 0 0.0000
5 Totl 2968184.968 100.00 215 13805.5115
------------------------------------------------------
trend_sRT_all_ANOVA =
labels: [4x5 char]
means: [734.2176 679.6898 655.1343 594.0093 583.6713 543.3009 566.7315 533.5093]
SS: [1.3869e+06 9.5664e+05 6.2461e+05 0 2.9682e+06]
MS: [5.3343e+04 1.3666e+05 3.4319e+03 0 1.3806e+04]
df: [26 7 182 0 215]
MS_err: 3.4319e+03
df_err: 182
F_omnibus: 39.8210
p_omnibus: 0
omega2_omnibus: 0.5189
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -178.6515
SS_linear: 8.6174e+05
F_linear: 251.0942
p_linear: 0
omega2_linear: 0.8224
SS_failure_linear: 9.4903e+04
df_failure_linear: 6
MS_failure_linear: 1.5817e+04
F_failure_linear: 4.6088
p_failure_linear: 2.2057e-04
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: 49.1303
SS_quad: 6.5172e+04
F_quad: 18.9898
p_quad: 2.1929e-05
omega2_quad: 0.2499
Linear (and quadratic) trend analysis of mean nondecision time Ter
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test Ter27x8=B(1).ter(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(v2_27x8(:),[sbj(:) period(:)],'SP') ; trend_Ter_ANOVA.labels = lbl' ; trend_Ter_ANOVA.means = mean(Ter27x8) ; trend_Ter_ANOVA.SS = SS' ; trend_Ter_ANOVA.MS = MS' ; trend_Ter_ANOVA.df = df' ; trend_Ter_ANOVA.MS_err = MS(3) ; trend_Ter_ANOVA.df_err = df(3) ; trend_Ter_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_Ter_ANOVA.p_omnibus = 1-fcdf(trend_Ter_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_Ter_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_Ter_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_Ter_ANOVA.linear_contrast_coefs = lin_coef ; trend_Ter_ANOVA.psi_hat_linear = trend_Ter_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_Ter_ANOVA.SS_linear = N_sbj * trend_Ter_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_Ter_ANOVA.F_linear = trend_Ter_ANOVA.SS_linear / trend_Ter_ANOVA.MS_err ; trend_Ter_ANOVA.p_linear = 1-fcdf(trend_Ter_ANOVA.F_linear,1,trend_Ter_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_Ter_ANOVA.F_linear-1) ; trend_Ter_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_Ter_ANOVA.SS_failure_linear = trend_Ter_ANOVA.SS(2) - trend_Ter_ANOVA.SS_linear ; trend_Ter_ANOVA.df_failure_linear = trend_Ter_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_Ter_ANOVA.MS_failure_linear = trend_Ter_ANOVA.SS_failure_linear / trend_Ter_ANOVA.df_failure_linear ; trend_Ter_ANOVA.F_failure_linear = trend_Ter_ANOVA.MS_failure_linear / trend_Ter_ANOVA.MS_err ; trend_Ter_ANOVA.p_failure_linear = 1-fcdf(trend_Ter_ANOVA.F_failure_linear,... trend_Ter_ANOVA.df_failure_linear, trend_Ter_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_Ter_ANOVA.quad_contrast_coefs = quad_coef ; trend_Ter_ANOVA.psi_hat_quad = trend_Ter_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_Ter_ANOVA.SS_quad = N_sbj * trend_Ter_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_Ter_ANOVA.F_quad = trend_Ter_ANOVA.SS_quad / trend_Ter_ANOVA.MS_err ; trend_Ter_ANOVA.p_quad = 1-fcdf(trend_Ter_ANOVA.F_quad,1,trend_Ter_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_Ter_ANOVA.F_quad-1) ; trend_Ter_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 1.022 45.18 26 0.0393
2 P 0.468 20.68 7 0.0668
3 SP 0.773 34.14 182 0.0042
4 err -0.000 -0.00 0 0.0000
5 Totl 2.262 100.00 215 0.0105
------------------------------------------------------
trend_Ter_ANOVA =
labels: [4x5 char]
means: [0.5100 0.4799 0.4776 0.4338 0.4334 0.4006 0.4164 0.4060]
SS: [1.0221 0.4679 0.7725 -7.1054e-15 2.2625]
MS: [0.0393 0.0668 0.0042 0 0.0105]
df: [26 7 182 0 215]
MS_err: 0.0042
df_err: 182
F_omnibus: 15.7463
p_omnibus: 4.4409e-16
omega2_omnibus: 0.2906
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -0.0985
SS_linear: 0.2622
F_linear: 61.7704
p_linear: 3.2652e-13
omega2_linear: 0.5295
SS_failure_linear: 0.2057
df_failure_linear: 6
MS_failure_linear: 0.0343
F_failure_linear: 8.0756
p_failure_linear: 9.6276e-08
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: 0.0261
SS_quad: 0.0183
F_quad: 4.3200
p_quad: 0.0391
omega2_quad: 0.0579
Linear (and quadratic) trend analysis of nondecision-time range parameter st
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test st27x8=B(1).st(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(st27x8(:),[sbj(:) period(:)],'SP') ; trend_st_ANOVA.labels = lbl' ; trend_st_ANOVA.means = mean(st27x8) ; trend_st_ANOVA.SS = SS' ; trend_st_ANOVA.MS = MS' ; trend_st_ANOVA.df = df' ; trend_st_ANOVA.MS_err = MS(3) ; trend_st_ANOVA.df_err = df(3) ; trend_st_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_st_ANOVA.p_omnibus = 1-fcdf(trend_st_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_st_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_st_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_st_ANOVA.linear_contrast_coefs = lin_coef ; trend_st_ANOVA.psi_hat_linear = trend_st_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_st_ANOVA.SS_linear = N_sbj * trend_st_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_st_ANOVA.F_linear = trend_st_ANOVA.SS_linear / trend_st_ANOVA.MS_err ; trend_st_ANOVA.p_linear = 1-fcdf(trend_st_ANOVA.F_linear,1,trend_st_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_st_ANOVA.F_linear-1) ; trend_st_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_st_ANOVA.SS_failure_linear = trend_st_ANOVA.SS(2) - trend_st_ANOVA.SS_linear ; trend_st_ANOVA.df_failure_linear = trend_st_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_st_ANOVA.MS_failure_linear = trend_st_ANOVA.SS_failure_linear / trend_st_ANOVA.df_failure_linear ; trend_st_ANOVA.F_failure_linear = trend_st_ANOVA.MS_failure_linear / trend_st_ANOVA.MS_err ; trend_st_ANOVA.p_failure_linear = 1-fcdf(trend_st_ANOVA.F_failure_linear,... trend_st_ANOVA.df_failure_linear, trend_st_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_st_ANOVA.quad_contrast_coefs = quad_coef ; trend_st_ANOVA.psi_hat_quad = trend_st_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_st_ANOVA.SS_quad = N_sbj * trend_st_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_st_ANOVA.F_quad = trend_st_ANOVA.SS_quad / trend_st_ANOVA.MS_err ; trend_st_ANOVA.p_quad = 1-fcdf(trend_st_ANOVA.F_quad,1,trend_st_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_st_ANOVA.F_quad-1) ; trend_st_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 0.479 24.44 26 0.0184
2 P 0.758 38.73 7 0.1084
3 SP 0.721 36.83 182 0.0040
4 err 0.000 0.00 0 0.0000
5 Totl 1.959 100.00 215 0.0091
------------------------------------------------------
trend_st_ANOVA =
labels: [4x5 char]
means: [0.2827 0.2416 0.1946 0.1511 0.1416 0.1107 0.1158 0.1249]
SS: [0.4787 0.7585 0.7213 0 1.9585]
MS: [0.0184 0.1084 0.0040 0 0.0091]
df: [26 7 182 0 215]
MS_err: 0.0040
df_err: 182
F_omnibus: 27.3400
p_omnibus: 0
omega2_omnibus: 0.4225
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -0.1539
SS_linear: 0.6396
F_linear: 161.3929
p_linear: 0
omega2_linear: 0.7481
SS_failure_linear: 0.1188
df_failure_linear: 6
MS_failure_linear: 0.0198
F_failure_linear: 4.9978
p_failure_linear: 9.1616e-05
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: 0.0642
SS_quad: 0.1112
F_quad: 28.0477
p_quad: 3.3878e-07
omega2_quad: 0.3337
Linear (and quadratic) trend analysis of minimum nondecision time t1
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1).
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test t1_27x8=B(1).t1(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(t1_27x8(:),[sbj(:) period(:)],'SP') ; trend_t1_ANOVA.labels = lbl' ; trend_t1_ANOVA.means = mean(t1_27x8) ; trend_t1_ANOVA.SS = SS' ; trend_t1_ANOVA.MS = MS' ; trend_t1_ANOVA.df = df' ; trend_t1_ANOVA.MS_err = MS(3) ; trend_t1_ANOVA.df_err = df(3) ; trend_t1_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_t1_ANOVA.p_omnibus = 1-fcdf(trend_t1_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_t1_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_t1_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_t1_ANOVA.linear_contrast_coefs = lin_coef ; trend_t1_ANOVA.psi_hat_linear = trend_t1_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_t1_ANOVA.SS_linear = N_sbj * trend_t1_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_t1_ANOVA.F_linear = trend_t1_ANOVA.SS_linear / trend_t1_ANOVA.MS_err ; trend_t1_ANOVA.p_linear = 1-fcdf(trend_t1_ANOVA.F_linear,1,trend_t1_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_t1_ANOVA.F_linear-1) ; trend_t1_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_t1_ANOVA.SS_failure_linear = trend_t1_ANOVA.SS(2) - trend_t1_ANOVA.SS_linear ; trend_t1_ANOVA.df_failure_linear = trend_t1_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_t1_ANOVA.MS_failure_linear = trend_t1_ANOVA.SS_failure_linear / trend_t1_ANOVA.df_failure_linear ; trend_t1_ANOVA.F_failure_linear = trend_t1_ANOVA.MS_failure_linear / trend_t1_ANOVA.MS_err ; trend_t1_ANOVA.p_failure_linear = 1-fcdf(trend_t1_ANOVA.F_failure_linear,... trend_t1_ANOVA.df_failure_linear, trend_t1_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_t1_ANOVA.quad_contrast_coefs = quad_coef ; trend_t1_ANOVA.psi_hat_quad = trend_t1_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_t1_ANOVA.SS_quad = N_sbj * trend_t1_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_t1_ANOVA.F_quad = trend_t1_ANOVA.SS_quad / trend_t1_ANOVA.MS_err ; trend_t1_ANOVA.p_quad = 1-fcdf(trend_t1_ANOVA.F_quad,1,trend_t1_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_t1_ANOVA.F_quad-1) ; trend_t1_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1 % %% Within-sbj ANOVA for the minimum nondecision time t1 % % % % Verify that t1 doesn't change significantly. % % This is old (Aug 2010) % sbj=repmat((1:N_sbj)',1,N_periods_total) ; % IV % period=repmat(1:N_periods_total,N_sbj,1) ; % IV % % [SS, df, MS, lbl] = anova(B(1).t1(:),[sbj(:) period(:)],'SP') ; % % t1_ANOVA.labels = lbl' ; % t1_ANOVA.SS = SS' ; % t1_ANOVA.MS = MS' ; % t1_ANOVA.df = df' ; % t1_ANOVA.F = MS(2)/MS(3) ; % MS('P')/MS('SxP') % t1_ANOVA.p = 1-fcdf(t1_ANOVA.F,df(2),df(3)) % % clear SS df MS lbl
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 0.580 47.49 26 0.0223
2 P 0.027 2.18 7 0.0038
3 SP 0.615 50.32 182 0.0034
4 err 0.000 0.00 0 0.0000
5 Totl 1.222 100.00 215 0.0057
------------------------------------------------------
trend_t1_ANOVA =
labels: [4x5 char]
means: [0.3686 0.3591 0.3803 0.3582 0.3626 0.3453 0.3585 0.3435]
SS: [0.5803 0.0267 0.6148 0 1.2218]
MS: [0.0223 0.0038 0.0034 0 0.0057]
df: [26 7 182 0 215]
MS_err: 0.0034
df_err: 182
F_omnibus: 1.1276
p_omnibus: 0.3476
omega2_omnibus: 0.0035
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -0.0216
SS_linear: 0.0126
F_linear: 3.7234
p_linear: 0.0552
omega2_linear: 0.0480
SS_failure_linear: 0.0141
df_failure_linear: 6
MS_failure_linear: 0.0023
F_failure_linear: 0.6950
p_failure_linear: 0.6539
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: -0.0060
SS_quad: 9.7918e-04
F_quad: 0.2898
p_quad: 0.5910
omega2_quad: 0
Linear (and quadratic) trend analysis of maximum nondecision time t2
Added by Alex 2010-10-22. All diffusion parameters come from the saturated model (aka MODEL_1). 2010-10-22: The maximum nondecisioni time cannot be estimated reliably from data and will not be emphasized in the revised manuscript.
% % We are only looking at the training portion (periods 1-8) % sbj=repmat((1:N_sbj)',1,N_periods_train) ; % IV % period=repmat(1:N_periods_train,N_sbj,1) ; % IV % Run a standard ANOVA analsis on the first 8 periods % We only want the first 8 points -- the training prior to the MAE and post-test t2_27x8=B(1).t2(:,1:N_periods_train) ; % [N_sbj x N_periods], dependent var [SS, df, MS, lbl] = anova(t2_27x8(:),[sbj(:) period(:)],'SP') ; trend_t2_ANOVA.labels = lbl' ; trend_t2_ANOVA.means = mean(t2_27x8) ; trend_t2_ANOVA.SS = SS' ; trend_t2_ANOVA.MS = MS' ; trend_t2_ANOVA.df = df' ; trend_t2_ANOVA.MS_err = MS(3) ; trend_t2_ANOVA.df_err = df(3) ; trend_t2_ANOVA.F_omnibus = MS(2)/MS(3) ; % MS('P')/MS('SxP') trend_t2_ANOVA.p_omnibus = 1-fcdf(trend_t2_ANOVA.F_omnibus,df(2),df(3)) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % The partial omega squared estimates the *population* ratio % sigma^2_effect / (sigma^2_effect + sigma^2_error) (Eq.21.6), % where sigma^2_error is the error variance for the particular effect. F1 = max(0,trend_t2_ANOVA.F_omnibus-1) ; % Eq. 8.12 in KeppelWickens04 a1 = df(2)-1 ; trend_t2_ANOVA.omega2_omnibus = (a1*F1) / (a1*F1 + N_periods_train*N_sbj) ; % Calculate the linear contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient. The grand_mean.a is from the original 27 sbjs %psi_hat = sum(lin_coef .* grand_means.a(1:N_periods_train)) ; trend_t2_ANOVA.linear_contrast_coefs = lin_coef ; trend_t2_ANOVA.psi_hat_linear = trend_t2_ANOVA.means * lin_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison %SS_linear = (N_sbj*(psi_hat^2)) / sum((lin_coef.^2)) ; trend_t2_ANOVA.SS_linear = N_sbj * trend_t2_ANOVA.psi_hat_linear^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_t2_ANOVA.F_linear = trend_t2_ANOVA.SS_linear / trend_t2_ANOVA.MS_err ; trend_t2_ANOVA.p_linear = 1-fcdf(trend_t2_ANOVA.F_linear,1,trend_t2_ANOVA.df_err) ; % Calculate effect size -- partial omega squared (KeppelWickens04, Sec.8.2) % For a contrast, the partial omega squared estimates the variability of % the contrast relative to itself and the error rather than to all the % variability in the study (p.165). It is not affected by the size of any % contrasts that are orthogonal to the contrast in question. F1 = max(0,trend_t2_ANOVA.F_linear-1) ; trend_t2_ANOVA.omega2_linear = F1 / (F1+2*N_sbj) ; % Eq. 8.17 in KeppelWickens04 % We can evaluate the linear fit to see if it accounts for the variance seen % See Eq. 4.16 in Keppel & Wickens (2004, p. 82) trend_t2_ANOVA.SS_failure_linear = trend_t2_ANOVA.SS(2) - trend_t2_ANOVA.SS_linear ; trend_t2_ANOVA.df_failure_linear = trend_t2_ANOVA.df(2) - 1 ; % Linear trend has 1 df trend_t2_ANOVA.MS_failure_linear = trend_t2_ANOVA.SS_failure_linear / trend_t2_ANOVA.df_failure_linear ; trend_t2_ANOVA.F_failure_linear = trend_t2_ANOVA.MS_failure_linear / trend_t2_ANOVA.MS_err ; trend_t2_ANOVA.p_failure_linear = 1-fcdf(trend_t2_ANOVA.F_failure_linear,... trend_t2_ANOVA.df_failure_linear, trend_t2_ANOVA.df_err) ; % Calculate the quadratic contrast % Find psi-hat by summing over the products of each periods mean and its % corresponding coefficient %psi_hat_quad = sum(quad_coef .* grand_means.a(1:N_periods_train)) ; trend_t2_ANOVA.quad_contrast_coefs = quad_coef ; trend_t2_ANOVA.psi_hat_quad = trend_t2_ANOVA.means * quad_coef' ; % Now we use equation 4.5 (pg 69) to complete the comparison trend_t2_ANOVA.SS_quad = N_sbj * trend_t2_ANOVA.psi_hat_quad^2 ; % sum(coef.^2)==1 % Finally, we test for significance using the MS_error from the ANOVA trend_t2_ANOVA.F_quad = trend_t2_ANOVA.SS_quad / trend_t2_ANOVA.MS_err ; trend_t2_ANOVA.p_quad = 1-fcdf(trend_t2_ANOVA.F_quad,1,trend_t2_ANOVA.df_err) ; % Calculate partial omega squared for the quadratic contrast. There must % be a better way to do this. As it is now, the two effect sizes (linear + % quadratic) can add up to more than 100%. F1 = max(0,trend_t2_ANOVA.F_quad-1) ; trend_t2_ANOVA.omega2_quad = F1 / (F1+2*N_sbj) % Eq. 8.17 in KeppelWickens04 clear SS df MS lbl F1 a1
Partitioning the sum of squares...
k Source SumSq eta2[%] df MeanSq
------------------------------------------------------
1 S 1.353 45.21 26 0.0521
2 P 0.951 31.77 7 0.1359
3 SP 0.689 23.02 182 0.0038
4 err 0.000 0.00 0 0.0000
5 Totl 2.994 100.00 215 0.0139
------------------------------------------------------
trend_t2_ANOVA =
labels: [4x5 char]
means: [0.6514 0.6007 0.5749 0.5094 0.5042 0.4559 0.4743 0.4684]
SS: [1.3535 0.9510 0.6891 3.5527e-14 2.9936]
MS: [0.0521 0.1359 0.0038 0 0.0139]
df: [26 7 182 0 215]
MS_err: 0.0038
df_err: 182
F_omnibus: 35.8782
p_omnibus: 0
omega2_omnibus: 0.4921
linear_contrast_coefs: [-0.5401 -0.3858 -0.2315 -0.0772 0.0772 0.2315 0.3858 0.5401]
psi_hat_linear: -0.1755
SS_linear: 0.8316
F_linear: 219.6310
p_linear: 0
omega2_linear: 0.8019
SS_failure_linear: 0.1193
df_failure_linear: 6
MS_failure_linear: 0.0199
F_failure_linear: 5.2527
p_failure_linear: 5.1529e-05
quad_contrast_coefs: [0.5401 0.0772 -0.2315 -0.3858 -0.3858 -0.2315 0.0772 0.5401]
psi_hat_quad: 0.0581
SS_quad: 0.0913
F_quad: 24.1056
p_quad: 2.0199e-06
omega2_quad: 0.2997
Bootstrap group-level Z-tests about boundary separation
These are superseded by the ANOVA above This is old (prior to Aug 2010)
% z-test for a_change111 := (a1-a11) to determine if the boundary separation % parameter has changed during the experiment a_change111 = a_gr(:,1) - a_gr(:,N_periods_total) ; describe(100*a_change111,'100*a_change111') ; a111_stats.mean = mean(a_change111) ; a111_stats.std = std(a_change111) ; a111_stats.z = a111_stats.mean ./ a111_stats.std ; a111_stats.onetailed_p = 1 - normcdf(a111_stats.z) % z-test for a_change18 := (a1-a8) to determine if the boundary separation % parameter has changed during the training phase only a_change18 = a_gr(:,1) - a_gr(:,N_periods_train) ; describe(100*a_change18,'100*a_change18') ; a18_stats.mean = mean(a_change18) ; a18_stats.std = std(a_change18) ; a18_stats.z = a18_stats.mean ./ a18_stats.std ; a18_stats.onetailed_p = 1 - normcdf(a18_stats.z)
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
0.515 0.812 -2.39 -0.04 0.50 1.08 2.77 100*a_change111
a111_stats =
mean: 0.0052
std: 0.0081
z: 0.6342
onetailed_p: 0.2630
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
1.262 0.791 -1.73 0.72 1.25 1.80 3.44 100*a_change18
a18_stats =
mean: 0.0126
std: 0.0079
z: 1.5947
onetailed_p: 0.0554
Bootstrap group-level Z-tests about nondecision times
These are superseded by the ANOVA above This is old (prior to Aug 2010)
% z-test for t1_change111 := (t1(1)-t1(11)) to determine if the minimum % non-decision time has changed during the experiment t1_change111 = t1_gr(:,1) - t1_gr(:,N_periods_total) ; describe(t1_change111,'t1_change111') ; t1_stats.mean = mean(t1_change111) ; t1_stats.std = std(t1_change111) ; t1_stats.z = t1_stats.mean ./ t1_stats.std ; t1_stats.onetailed_p = 1 - normcdf(t1_stats.z) % z-test for t1_change18 := (t1(1)-t1(8)) to determine if the minimum % non-decision time has changed during the training phase t1_change18 = t1_gr(:,1) - t1_gr(:,N_periods_train) ; describe(t1_change18,'t1_change18') ; t1_18_stats.mean = mean(t1_change18) ; t1_18_stats.std = std(t1_change18) ; t1_18_stats.z = t1_18_stats.mean ./ t1_18_stats.std ; t1_18_stats.onetailed_p = 1 - normcdf(t1_18_stats.z)
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
0.010 0.027 -0.08 -0.01 0.01 0.03 0.10 t1_change111
t1_stats =
mean: 0.0096
std: 0.0273
z: 0.3531
onetailed_p: 0.3620
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
0.025 0.026 -0.06 0.01 0.02 0.04 0.11 t1_change18
t1_18_stats =
mean: 0.0249
std: 0.0255
z: 0.9738
onetailed_p: 0.1651
Descriptive statistics of the drop of the maximum nondecision time t2
Calculated across 1000 bootstrap samples of the group averages t2_gr This is old (prior to Aug 2010). 2010-10-20: The maximum nondecisioni time cannot be estimated reliably from data and will not be emphasized in the revised manuscript.
t2_drop18_gr = 1000.*(t2_gr(:,1)-t2_gr(:,N_periods_train)) ; % milliseconds describe(t2_drop18_gr,'t2=Ter+st/2, drop period 1 - 8') norminv(.90)*std(t2_drop18_gr) t2_drop111_gr = 1000.*(t2_gr(:,1)-t2_gr(:,N_periods_total)) ; % milliseconds describe(t2_drop111_gr,'t2=Ter+st/2, drop period 1 - 11') norminv(.90)*std(t2_drop111_gr) % In the text, this was simplified to delta_Tmax = 185+-25 ms
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
182.578 17.548 122.67 171.05 183.11 194.04 231.33 t2=Ter+st/2, drop period 1 - 8
ans =
22.4892
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
192.137 21.118 132.15 177.77 192.12 206.82 249.35 t2=Ter+st/2, drop period 1 - 11
ans =
27.0633
Descriptive statistics of the drop of the mean RT
This is old (prior to Aug 2010).
RT_all_drop18_gr = (RT_all_gr(:,1)-RT_all_gr(:,N_periods_train)) ; % milliseconds describe(RT_all_drop18_gr,'RT_all, drop period 1 - 8') norminv(.90)*std(RT_all_drop18_gr) RT_all_drop111_gr = (RT_all_gr(:,1)-RT_all_gr(:,N_periods_total)) ; % milliseconds describe(RT_all_drop111_gr,'RT_all, drop period 1 - 11') norminv(.90)*std(RT_all_drop111_gr)
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
199.974 23.013 121.15 184.59 200.19 213.83 282.69 RT_all, drop period 1 - 8
ans =
29.4923
Mean Std.dev Min Q25 Median Q75 Max
------------------------------------------------------------
161.633 23.524 73.24 145.62 161.79 176.29 255.34 RT_all, drop period 1 - 11
ans =
30.1473